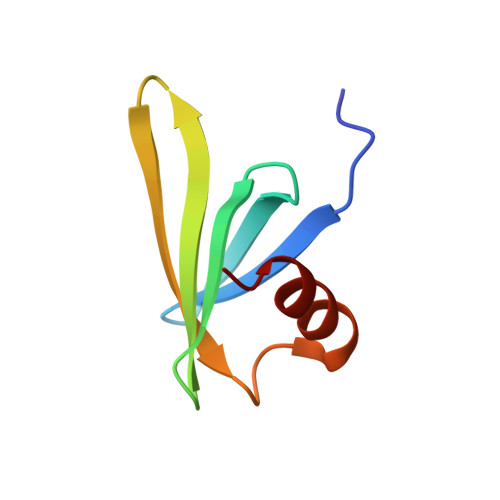
Genetically encoded live-cell sensor for tyrosinated microtubules.
Kesarwani, S., Lama, P., Chandra, A., Reddy, P.P., Jijumon, A.S., Bodakuntla, S., Rao, B.M., Janke, C., Das, R., Sirajuddin, M.(2020) J Cell Biol 219
- PubMed: 32886100
- DOI: https://doi.org/10.1083/jcb.201912107
- Primary Citation of Related Structures:
7C1M - PubMed Abstract:
Microtubule cytoskeleton exists in various biochemical forms in different cells due to tubulin posttranslational modifications (PTMs). Tubulin PTMs are known to affect microtubule stability, dynamics, and interaction with MAPs and motors in a specific manner, widely known as tubulin code hypothesis. At present, there exists no tool that can specifically mark tubulin PTMs in living cells, thus severely limiting our understanding of their dynamics and cellular functions. Using a yeast display library, we identified a binder against terminal tyrosine of α-tubulin, a unique PTM site. Extensive characterization validates the robustness and nonperturbing nature of our binder as tyrosination sensor, a live-cell tubulin nanobody specific towards tyrosinated microtubules. Using this sensor, we followed nocodazole-, colchicine-, and vincristine-induced depolymerization events of tyrosinated microtubules in real time and found each distinctly perturbs the microtubule polymer. Together, our work describes a novel tyrosination sensor and its potential applications to study the dynamics of microtubule and their PTM processes in living cells.
- Centre for Cardiovascular Biology and Diseases, Institute for Stem Cell Science and Regenerative Medicine, Gandhi Krishi Vigyan Kendra Campus, Bangalore, India.
Organizational Affiliation: