Structural insights into the histidine-containing phospho-transfer protein and receiver domain of sensor histidine kinase suggest a complex model in the two-component regulatory system in Pseudomonas aeruginosa
Chen, S.K., Guan, H.H., Wu, P.H., Lin, L.T., Wu, M.C., Chang, H.Y., Chen, N.C., Lin, C.C., Chuankhayan, P., Huang, Y.C., Lin, P.J., Chen, C.J.(2020) IUCrJ 7: 934-948
- PubMed: 32939285
- DOI: https://doi.org/10.1107/S2052252520009665
- Primary Citation of Related Structures:
7C1I, 7C1J, 7CFW - PubMed Abstract:
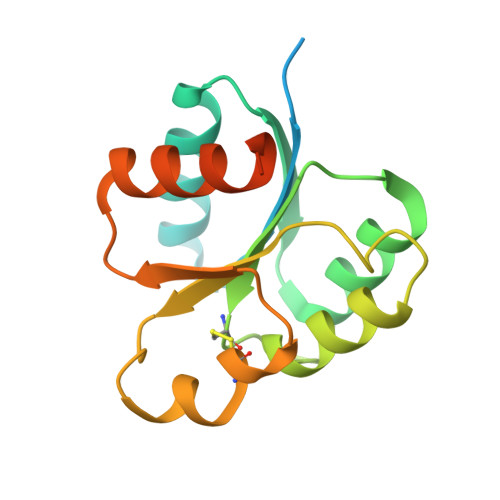
In Pseudomonas aeruginosa , an important opportunistic pathogen that causes numerous acute and chronic infections, the hybrid two-component system (TCS) regulates the swarming ability and biofilm formation with a multistep phospho-relay, and consists of hybrid-sensor histidine kinase (HK), histidine-containing phospho-transfer protein (Hpt) and response regulator (RR). In this work, two crystal structures of HptB and the receiver domain of HK PA1611 (PA1611REC) of P. aeruginosa have been determined in order to elucidate their interactions for the transfer of the phospho-ryl group. The structure of HptB folds into an elongated four-helix bundle - helices α2, α3, α4 and α5, covered by the short N-terminal helix α1. The imidazole side chain of the conserved active-site histidine residue His57, located near the middle of helix α3, protrudes from the bundle and is exposed to solvent. The structure of PA1611REC possesses a conventional (β/α) 5 topology with five-stranded parallel β-sheets folded in the central region, surrounded by five α-helices. The divalent Mg 2+ ion is located in the negatively charged active-site cleft and interacts with Asp522, Asp565 and Arg567. The HptB-PA1611REC complex is further modeled to analyze the binding surface and interactions between the two proteins. The model shows a shape complementarity between the convex surface of PA1611REC and the kidney-shaped HptB with fewer residues and a different network involved in interactions compared with other TCS complexes, such as SLN1-R1/YPD1 from Saccharomyces cerevisiae and AHK5 RD /AHP1 from Arabidopsis thaliana . These structural results provide a better understanding of the TCS in P. aeruginosa and could potentially lead to the discovery of a new treatment for infection.
- Department of Biotechnology and Bioindustry Sciences, National Cheng Kung University, Tainan City 701, Taiwan.
Organizational Affiliation: