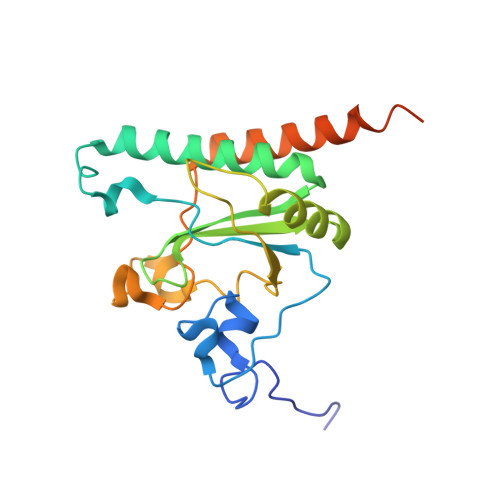
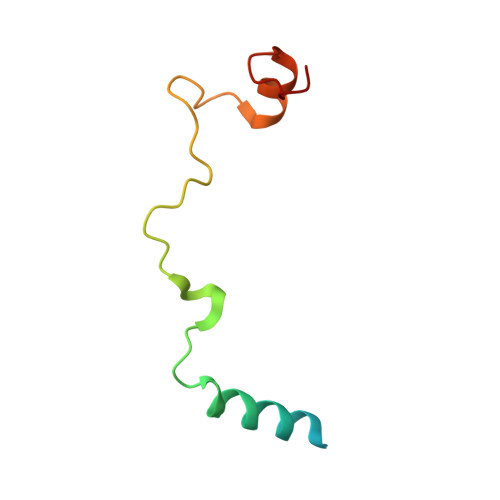
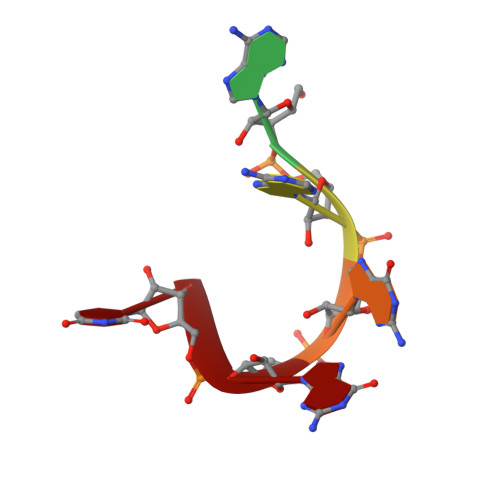
Elucidation of the aberrant 3' splice site selection by cancer-associated mutations on the U2AF1.
Yoshida, H., Park, S.Y., Sakashita, G., Nariai, Y., Kuwasako, K., Muto, Y., Urano, T., Obayashi, E.(2020) Nat Commun 11: 4744-4744
- PubMed: 32958768
- DOI: https://doi.org/10.1038/s41467-020-18559-6
- Primary Citation of Related Structures:
7C06, 7C07, 7C08 - PubMed Abstract:
The accurate exclusion of introns by RNA splicing is critical for the production of mature mRNA. U2AF1 binds specifically to the 3´ splice site, which includes an essential AG dinucleotide. Even a single amino acid mutation of U2AF1 can cause serious disease such as certain cancers or myelodysplastic syndromes. Here, we describe the first crystal structures of wild-type and pathogenic mutant U2AF1 complexed with target RNA, revealing the mechanism of 3´ splice site selection, and how aberrant splicing results from clinically important mutations. Unexpected features of this mechanism may assist the future development of new treatments against diseases caused by splicing errors.
- Graduate School of Medical Life Science, Yokohama City University, 1-7-29 Suehiro-cho, Tsurumi-ku, Yokohama, 230-0045, Japan.
Organizational Affiliation: