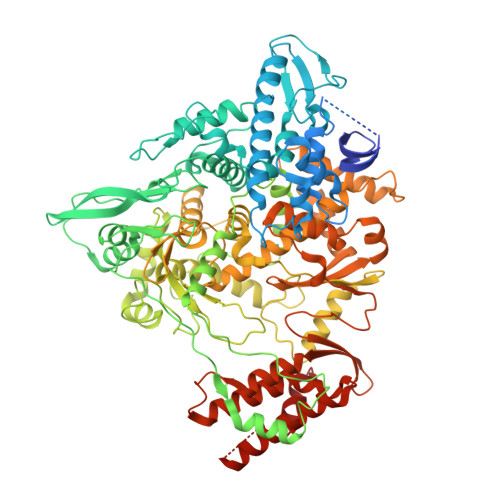
Structural and Biochemical Characterization of the nsp12-nsp7-nsp8 Core Polymerase Complex from SARS-CoV-2.
Peng, Q., Peng, R., Yuan, B., Zhao, J., Wang, M., Wang, X., Wang, Q., Sun, Y., Fan, Z., Qi, J., Gao, G.F., Shi, Y.(2020) Cell Rep 31: 107774-107774
- PubMed: 32531208
- DOI: https://doi.org/10.1016/j.celrep.2020.107774
- Primary Citation of Related Structures:
7BW4 - PubMed Abstract:
The ongoing global pandemic of coronavirus disease 2019 (COVID-19) has caused a huge number of human deaths. Currently, there are no specific drugs or vaccines available for this virus (SARS-CoV-2). The viral polymerase is a promising antiviral target. Here, we describe the near-atomic-resolution structure of the SARS-CoV-2 polymerase complex consisting of the nsp12 catalytic subunit and nsp7-nsp8 cofactors. This structure highly resembles the counterpart of SARS-CoV with conserved motifs for all viral RNA-dependent RNA polymerases and suggests a mechanism of activation by cofactors. Biochemical studies reveal reduced activity of the core polymerase complex and lower thermostability of individual subunits of SARS-CoV-2 compared with SARS-CoV. These findings provide important insights into RNA synthesis by coronavirus polymerase and indicate adaptation of SARS-CoV-2 toward humans with a relatively lower body temperature than the natural bat hosts.
- CAS Key Laboratory of Pathogenic Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China.
Organizational Affiliation: