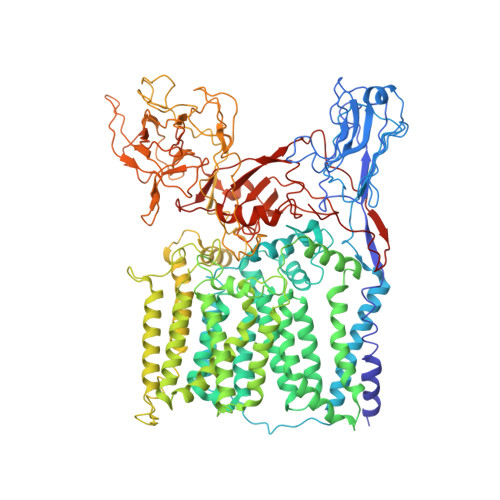
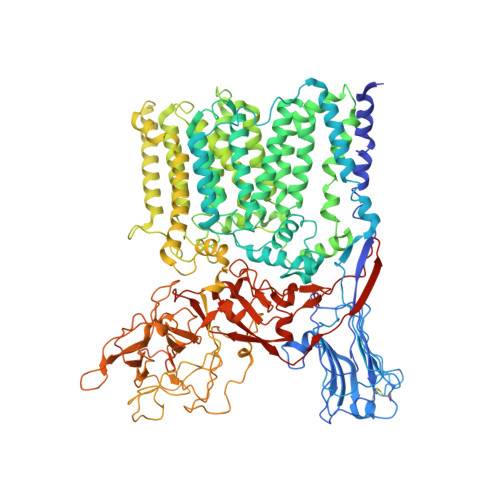
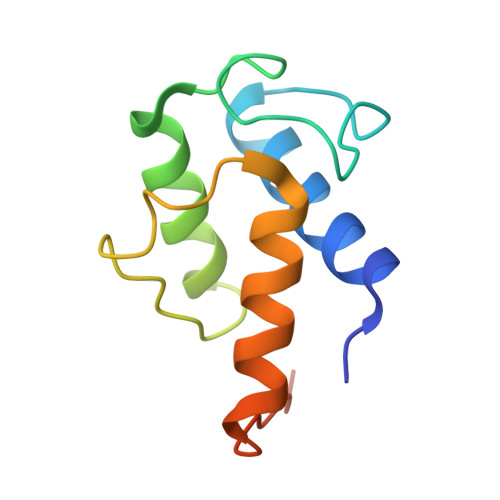
Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Zhang, L., Zhao, Y., Gao, Y., Wu, L., Gao, R., Zhang, Q., Wang, Y., Wu, C., Wu, F., Gurcha, S.S., Veerapen, N., Batt, S.M., Zhao, W., Qin, L., Yang, X., Wang, M., Zhu, Y., Zhang, B., Bi, L., Zhang, X., Yang, H., Guddat, L.W., Xu, W., Wang, Q., Li, J., Besra, G.S., Rao, Z.(2020) Science 368: 1211-1219
- PubMed: 32327601
- DOI: https://doi.org/10.1126/science.aba9102
- Primary Citation of Related Structures:
7BVC, 7BVE, 7BVF, 7BVG, 7BVH - PubMed Abstract:
The arabinosyltransferases EmbA, EmbB, and EmbC are involved in Mycobacterium tuberculosis cell wall synthesis and are recognized as targets for the anti-tuberculosis drug ethambutol. In this study, we determined cryo-electron microscopy and x-ray crystal structures of mycobacterial EmbA-EmbB and EmbC-EmbC complexes in the presence of their glycosyl donor and acceptor substrates and with ethambutol. These structures show how the donor and acceptor substrates bind in the active site and how ethambutol inhibits arabinosyltransferases by binding to the same site as both substrates in EmbB and EmbC. Most drug-resistant mutations are located near the ethambutol binding site. Collectively, our work provides a structural basis for understanding the biochemical function and inhibition of arabinosyltransferases and the development of new anti-tuberculosis agents.
- Shanghai Institute for Advanced Immunochemical Studies, iHuman Institute, School of Life Science and Technology, ShanghaiTech University, Shanghai 201210, China.
Organizational Affiliation: