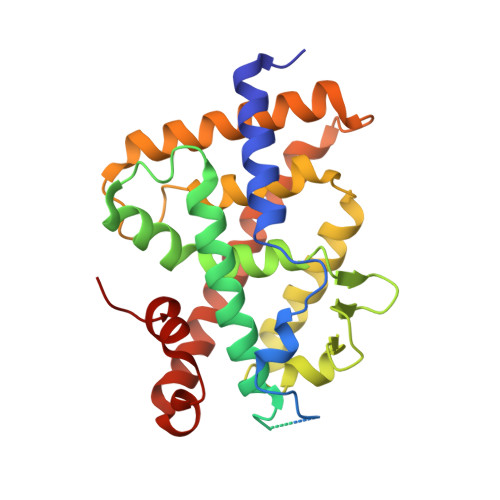
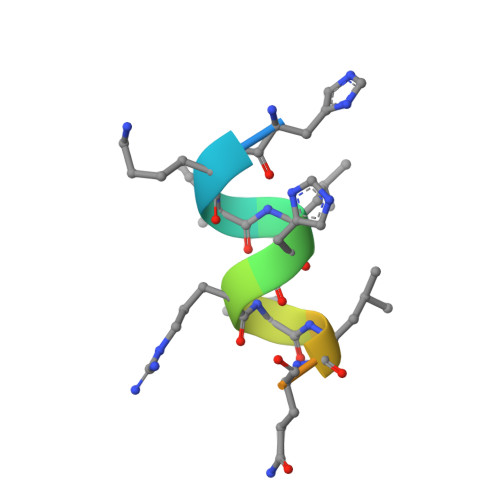
Lithocholic acid-based design of noncalcemic vitamin D receptor agonists.
Gaikwad, S., Gonzalez, C.M., Vilarino, D., Lasanta, G., Villaverde, C., Mourino, A., Verlinden, L., Verstuyf, A., Peluso-Iltis, C., Rochel, N., Berkowska, K., Marcinkowska, E.(2021) Bioorg Chem 111: 104878-104878
- PubMed: 33853023
- DOI: https://doi.org/10.1016/j.bioorg.2021.104878
- Primary Citation of Related Structures:
7BO6 - PubMed Abstract:
The hypercalcemic effects of the hormone 1α,25-dihydroxyvitamin D 3 (calcitriol) and most of known vitamin D metabolites and analogs call for the development of non secosteroidal vitamin D receptor (VDR) ligands as new selective and noncalcemic agonists for treatment of hyperproliferative diseases. We report on the in silico design and stereoselective synthesis of six lithocholic acid derivatives as well as on the calcemic activity of a potent LCA derivative and its crystallographic structure in complex with zVDR LBD. The low calcemic activity of this compound in comparison with the native hormone makes it of potential therapeutic value. Structure-function relationships provide the basis for the development of even more potent and selective lithocholic acid-based VDR ligands.
- Departamento de Química Orgánica, Laboratorio de Investigación Ignacio Ribas, Universidad de Santiago de Compostela, Avda das Ciencias s/n, 15782 Santiago de Compostela, Spain.
Organizational Affiliation: