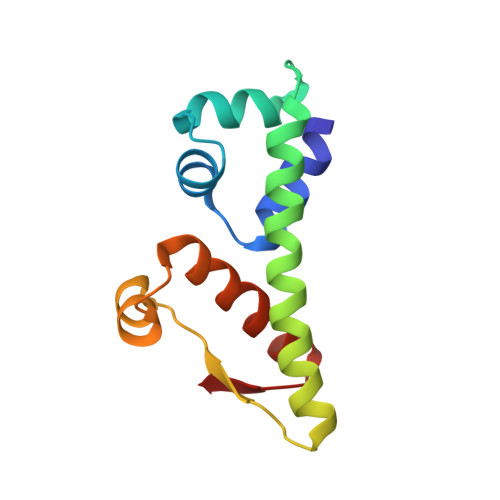
Crystal structure of the anti-CRISPR repressor Aca2.
Usher, B., Birkholz, N., Beck, I.N., Fagerlund, R.D., Jackson, S.A., Fineran, P.C., Blower, T.R.(2021) J Struct Biol 213: 107752-107752
- PubMed: 34116143
- DOI: https://doi.org/10.1016/j.jsb.2021.107752
- Primary Citation of Related Structures:
7B5J, 7ZGE - PubMed Abstract:
Bacteria use adaptive CRISPR-Cas immune mechanisms to protect from invasion by bacteriophages and other mobile genetic elements. In response, bacteriophages and mobile genetic elements have co-evolved anti-CRISPR proteins to inhibit the bacterial defense. We and others have previously shown that anti-CRISPR associated (Aca) proteins can regulate this anti-CRISPR counter-attack. Here, we report the first structure of an Aca protein, the Aca2 DNA-binding transcriptional autorepressor from Pectobacterium carotovorum bacteriophage ZF40, determined to 1.34 Å. Aca2 presents a conserved N-terminal helix-turn-helix DNA-binding domain and a previously uncharacterized C-terminal dimerization domain. Dimerization positions the Aca2 recognition helices for insertion into the major grooves of target DNA, supporting its role in regulating anti-CRISPRs. Furthermore, database comparisons identified uncharacterized Aca2 structural homologs in pathogenic bacteria, suggesting that Aca2 represents the first characterized member of a more widespread family of transcriptional regulators.
- Department of Biosciences, Durham University, Stockton Road, Durham DH1 3LE, UK.
Organizational Affiliation: