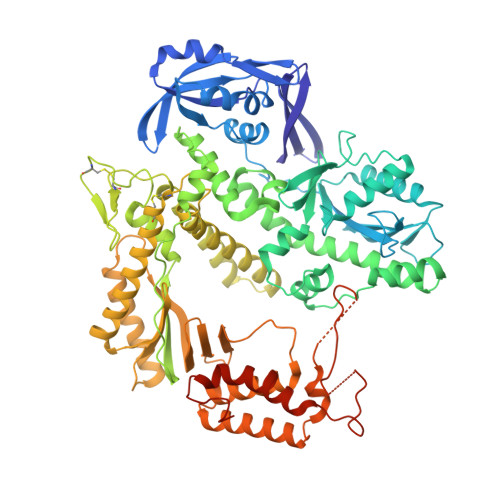
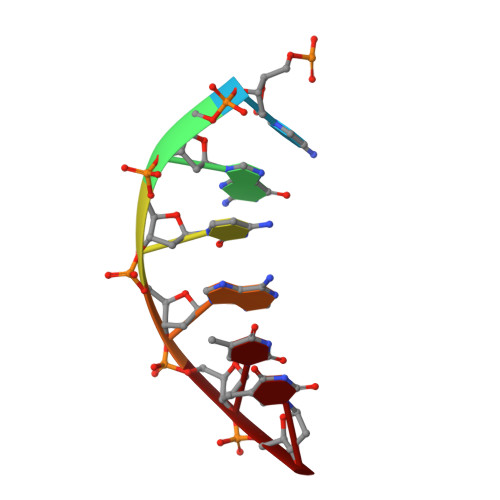
Structural Studies of HNA Substrate Specificity in Mutants of an Archaeal DNA Polymerase Obtained by Directed Evolution.
Samson, C., Legrand, P., Tekpinar, M., Rozenski, J., Abramov, M., Holliger, P., Pinheiro, V.B., Herdwijn, P., Delarue, M.(2020) Biomolecules 10
- PubMed: 33302546
- DOI: https://doi.org/10.3390/biom10121647
- Primary Citation of Related Structures:
7B06, 7B07, 7B08, 7B0F, 7B0G, 7B0H - PubMed Abstract:
Archaeal DNA polymerases from the B-family (polB) have found essential applications in biotechnology. In addition, some of their variants can accept a wide range of modified nucleotides or xenobiotic nucleotides, such as 1,5-anhydrohexitol nucleic acid (HNA), which has the unique ability to selectively cross-pair with DNA and RNA. This capacity is essential to allow the transmission of information between different chemistries of nucleic acid molecules. Variants of the archaeal polymerase from Thermococcus gorgonarius, TgoT, that can either generate HNA from DNA (TgoT_6G12) or DNA from HNA (TgoT_RT521) have been previously identified. To understand how DNA and HNA are recognized and selected by these two laboratory-evolved polymerases, we report six X-ray structures of these variants, as well as an in silico model of a ternary complex with HNA. Structural comparisons of the apo form of TgoT_6G12 together with its binary and ternary complexes with a DNA duplex highlight an ensemble of interactions and conformational changes required to promote DNA or HNA synthesis. MD simulations of the ternary complex suggest that the HNA-DNA hybrid duplex remains stable in the A-DNA helical form and help explain the presence of mutations in regions that would normally not be in contact with the DNA if it were not in the A-helical form. One complex with two incorporated HNA nucleotides is surprisingly found in a one nucleotide-backtracked form, which is new for a DNA polymerase. This information can be used for engineering a new generation of more efficient HNA polymerase variants.
- Unit of Structural Dynamics of Biological Macromolecules, UMR 3528 du CNRS, Institut Pasteur, 25-28 rue du Dr Roux, 75015 Paris, France.
Organizational Affiliation: