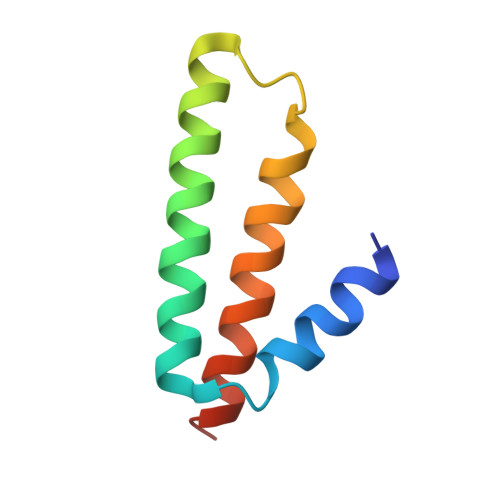
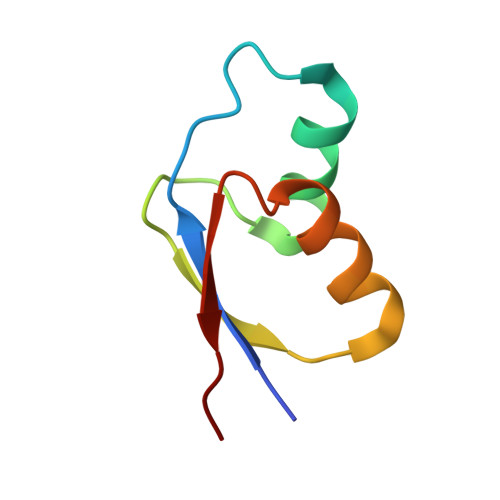
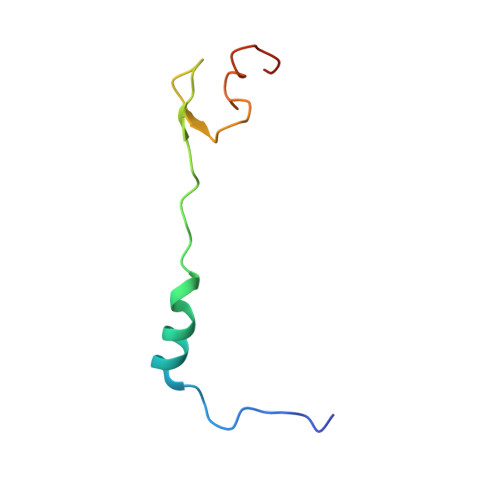
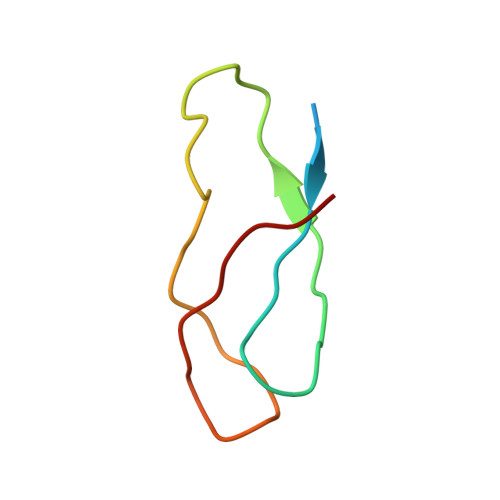
OHX Query on OHX Download Ideal Coordinates CCD File AEA [auth 23SA]
AEA [auth 23SA],
osmium (III) hexammine 12 N6 OsK Query on K Download Ideal Coordinates CCD File AAA [auth 23SA]
AAA [auth 23SA],
POTASSIUM ION MG Query on MG Download Ideal Coordinates CCD File AAB [auth 23SB]
AAB [auth 23SB],
MAGNESIUM ION