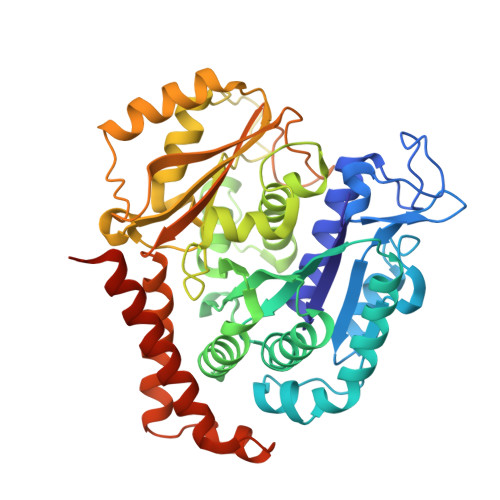
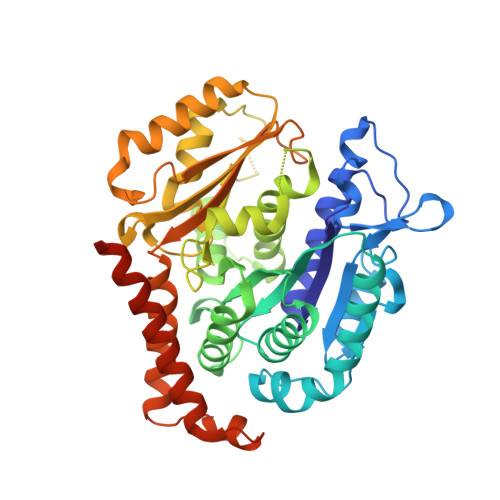
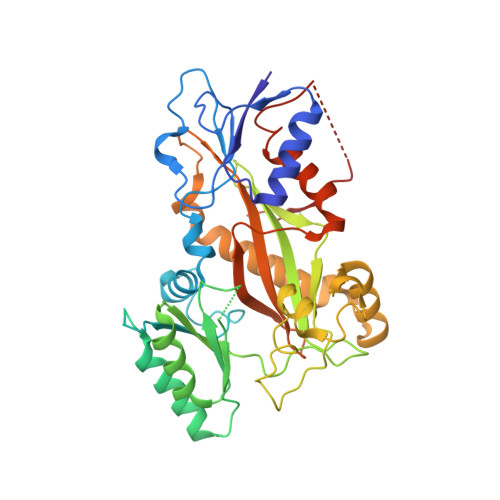
1,3-Benzodioxole-Modified Noscapine Analogues: Synthesis, Antiproliferative Activity, and Tubulin-Bound Structure.
Yong, C., Devine, S.M., Abel, A.C., Tomlins, S.D., Muthiah, D., Gao, X., Callaghan, R., Steinmetz, M.O., Prota, A.E., Capuano, B., Scammells, P.J.(2021) ChemMedChem 16: 2882-2894
- PubMed: 34159741
- DOI: https://doi.org/10.1002/cmdc.202100363
- Primary Citation of Related Structures:
7AU5 - PubMed Abstract:
Since the revelation of noscapine's weak anti-mitotic activity, extensive research has been conducted over the past two decades, with the goal of discovering noscapine derivatives with improved potency. To date, noscapine has been explored at the 1, 7, 6', and 9'-positions, though the 1,3-benzodioxole motif in the noscapine scaffold that remains unexplored. The present investigation describes the design, synthesis and pharmacological evaluation of noscapine analogues consisting of modifications to the 1,3-benzodioxole moiety. This includes expansion of the dioxolane ring and inclusion of metabolically robust deuterium and fluorine atoms. Favourable structural modifications were subsequently incorporated into multi-functionalised noscapine derivatives that also possessed modifications previously shown to promote anti-proliferative activity in the 1-, 6'- and 9'-positions. Our research efforts afforded the deuterated noscapine derivative 14 e and the dioxino-containing analogue 20 as potent cytotoxic agents with EC 50 values of 1.50 and 0.73 μM, respectively, against breast cancer (MCF-7) cells. Compound 20 also exhibited EC 50 values of <2 μM against melanoma, non-small cell lung carcinoma, and cancers of the brain, kidney and breast in an NCI screen. Furthermore, compounds 14 e and 20 inhibit tubulin polymerisation and are not vulnerable to the overexpression of resistance conferring P-gp efflux pumps in drug-resistant breast cancer cells (NCI ADR/RES ). We also conducted X-ray crystallography studies that yielded the high-resolution structure of 14 e bound to tubulin. Our structural analysis revealed the key interactions between this noscapinoid and tubulin and will assist with the future design of noscapine derivatives with improved properties.
- Medicinal Chemistry, Monash Institute of Pharmaceutical Sciences, Monash University, Parkville, VIC, 3052, Australia.
Organizational Affiliation: