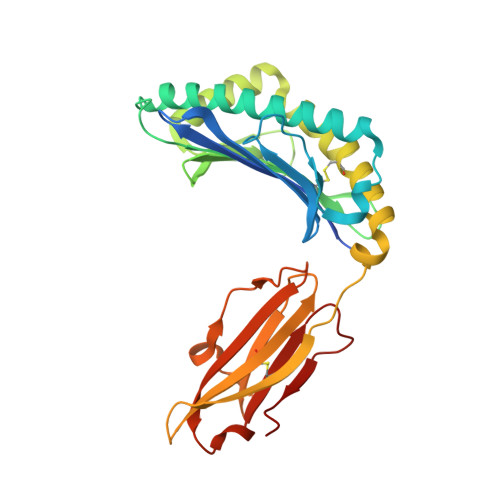
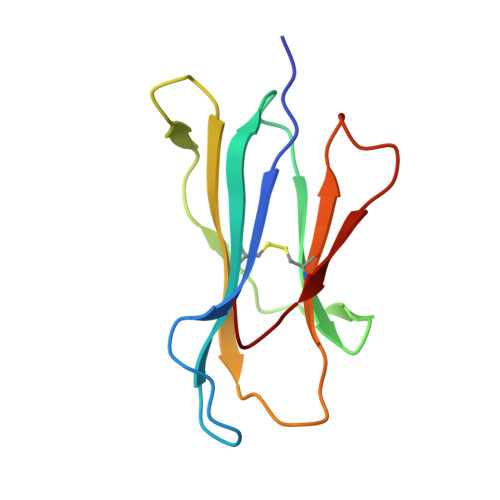
Exchange catalysis by tapasin exploits conserved and allele-specific features of MHC-I molecules.
Lan, H., Abualrous, E.T., Sticht, J., Fernandez, L.M.A., Werk, T., Weise, C., Ballaschk, M., Schmieder, P., Loll, B., Freund, C.(2021) Nat Commun 12: 4236-4236
- PubMed: 34244493
- DOI: https://doi.org/10.1038/s41467-021-24401-4
- Primary Citation of Related Structures:
7ALO - PubMed Abstract:
The repertoire of peptides presented by major histocompatibility complex class I (MHC-I) molecules on the cell surface is tailored by the ER-resident peptide loading complex (PLC), which contains the exchange catalyst tapasin. Tapasin stabilizes MHC-I molecules and promotes the formation of stable peptide-MHC-I (pMHC-I) complexes that serve as T cell antigens. Exchange of suboptimal by high-affinity ligands is catalyzed by tapasin, but the underlying mechanism is still elusive. Here we analyze the tapasin-induced changes in MHC-I dynamics, and find the catalyst to exploit two essential features of MHC-I. First, tapasin recognizes a conserved allosteric site underneath the α 2-1 -helix of MHC-I, 'loosening' the MHC-I F-pocket region that accomodates the C-terminus of the peptide. Second, the scoop loop 11-20 of tapasin relies on residue L18 to target the MHC-I F-pocket, enabling peptide exchange. Meanwhile, tapasin residue K16 plays an accessory role in catalysis of MHC-I allotypes bearing an acidic F-pocket. Thus, our results provide an explanation for the observed allele-specificity of catalyzed peptide exchange.
- Laboratory of Protein Biochemistry, Institute for Chemistry & Biochemistry, Freie Universität Berlin, Berlin, Germany.
Organizational Affiliation: