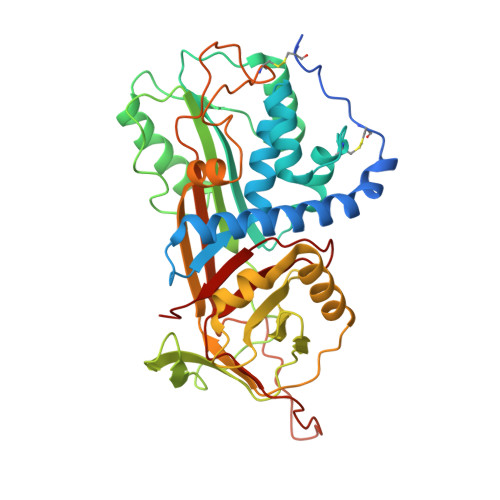
Molecular Basis for Bordetella pertussis Interference with Complement, Coagulation, Fibrinolytic, and Contact Activation Systems: the Cryo-EM Structure of the Vag8-C1 Inhibitor Complex.
Dhillon, A., Deme, J.C., Furlong, E., Roem, D., Jongerius, I., Johnson, S., Lea, S.M.(2021) mBio 12
- PubMed: 33758081
- DOI: https://doi.org/10.1128/mBio.02823-20
- Primary Citation of Related Structures:
7AKV - PubMed Abstract:
Complement, contact activation, coagulation, and fibrinolysis are serum protein cascades that need strict regulation to maintain human health. Serum glycoprotein, a C1 inhibitor (C1-INH), is a key regulator (inhibitor) of serine proteases of all the above-mentioned pathways. Recently, an autotransporter protein, virulence-associated gene 8 (Vag8), produced by the whooping cough pathogen, Bordetella pertussis , was shown to bind to C1-INH and interfere with its function. Here, we present the structure of the Vag8-C1-INH complex determined using cryo-electron microscopy at a 3.6-Å resolution. The structure shows a unique mechanism of C1-INH inhibition not employed by other pathogens, where Vag8 sequesters the reactive center loop of C1-INH, preventing its interaction with the target proteases. IMPORTANCE The structure of a 10-kDa protein complex is one of the smallest to be determined using cryo-electron microscopy at high resolution. The structure reveals that C1-INH is sequestered in an inactivated state by burial of the reactive center loop in Vag8. By so doing, the bacterium is able to simultaneously perturb the many pathways regulated by C1-INH. Virulence mechanisms such as the one described here assume more importance given the emerging evidence about dysregulation of contact activation, coagulation, and fibrinolysis leading to COVID-19 pneumonia.
- Sir William Dunn School of Pathology, Oxford, United Kingdom.
Organizational Affiliation: