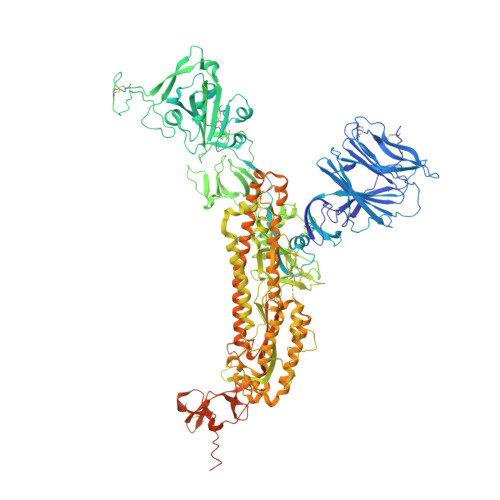
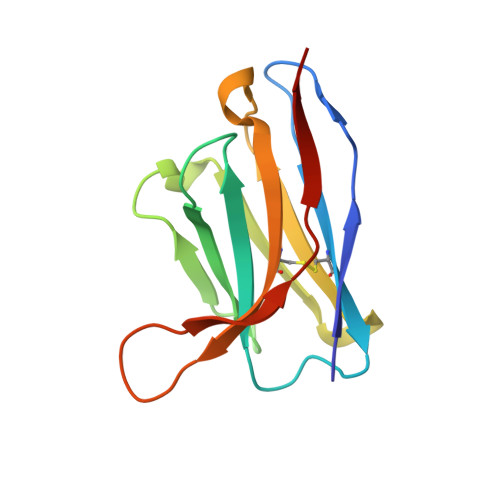
Structural insights into the cross-neutralization of SARS-CoV and SARS-CoV-2 by the human monoclonal antibody 47D11.
Fedry, J., Hurdiss, D.L., Wang, C., Li, W., Obal, G., Drulyte, I., Du, W., Howes, S.C., van Kuppeveld, F.J.M., Forster, F., Bosch, B.J.(2021) Sci Adv 7
- PubMed: 33958322
- DOI: https://doi.org/10.1126/sciadv.abf5632
- Primary Citation of Related Structures:
7AKD, 7AKJ - PubMed Abstract:
The emergence of SARS-CoV-2 antibody escape mutations highlights the urgent need for broadly neutralizing therapeutics. We previously identified a human monoclonal antibody, 47D11, capable of cross-neutralizing SARS-CoV-2 and SARS-CoV and protecting against the associated respiratory disease in an animal model. Here, we report cryo-EM structures of both trimeric spike ectodomains in complex with the 47D11 Fab. 47D11 binds to the closed receptor-binding domain, distal to the ACE2 binding site. The CDRL3 stabilizes the N343 glycan in an upright conformation, exposing a mutationally constrained hydrophobic pocket, into which the CDRH3 loop inserts two aromatic residues. 47D11 stabilizes a partially open conformation of the SARS-CoV-2 spike, suggesting that it could be used effectively in combination with other antibodies targeting the exposed receptor-binding motif. Together, these results reveal a cross-protective epitope on the SARS-CoV-2 spike and provide a structural roadmap for the development of 47D11 as a prophylactic or postexposure therapy for COVID-19.
- Cryo-Electron Microscopy, Bijvoet Center for Biomolecular Research, Department of Chemistry, Faculty of Science, Utrecht University, Padualaan 8, 3584 CH Utrecht, Netherlands.
Organizational Affiliation: