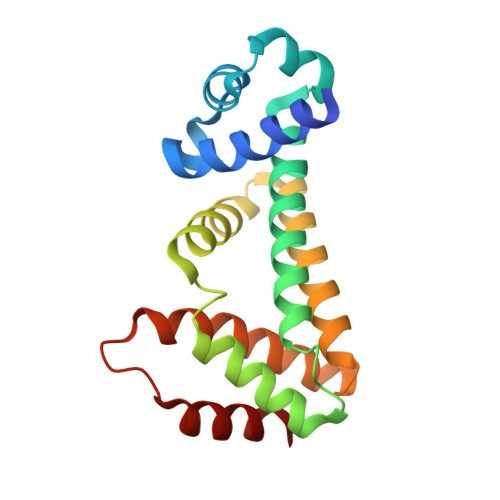
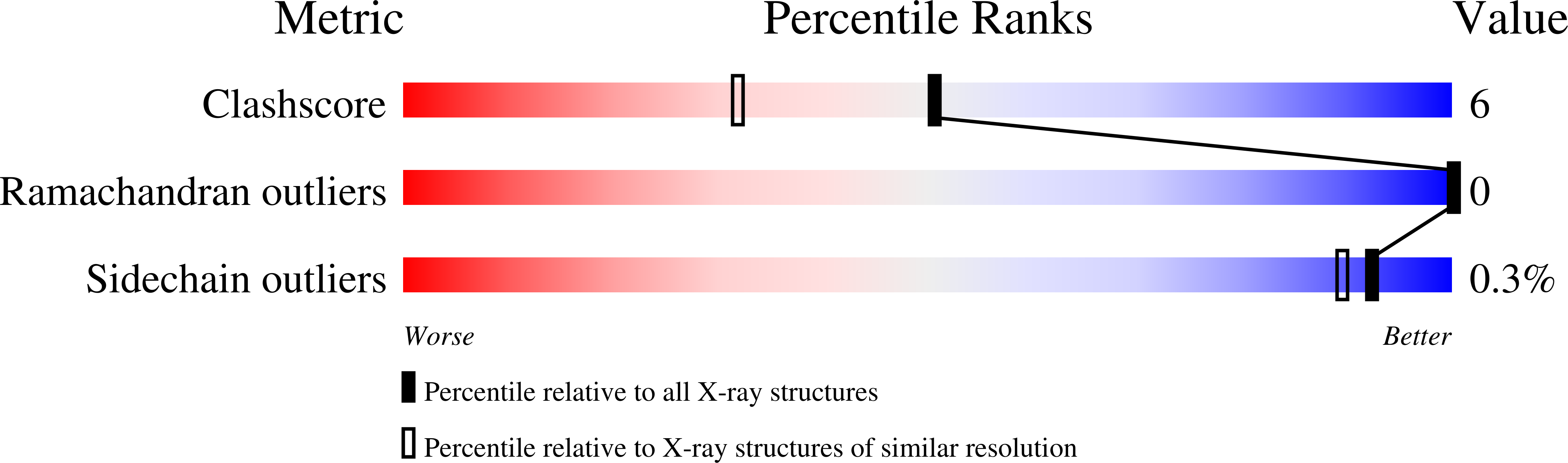Structures of the TetR-like transcription regulator RcdA alone and in complexes with ligands.
Pietrzyk-Brzezinska, A.J., Cociurovscaia, A.(2022) Proteins 90: 33-44
- PubMed: 34288132
- DOI: https://doi.org/10.1002/prot.26183
- Primary Citation of Related Structures:
7ACL, 7ACM, 7ACO - PubMed Abstract:
RcdA is a helix-turn-helix (HTH) transcriptional regulator belonging to the TetR family. The protein regulates the transcription of curlin subunit gene D, the master regulator of biofilm formation. Moreover, it was predicted that it might be involved in the regulation of up to 27 different genes. However, an effector of RcdA and the environmental conditions which trigger RcdA action remain unknown. Herein, we report the first crystal structures of RcdA in complexes with ligands, trimethylamine N-oxide (TMAO) and tris(hydroxymethyl)aminomethane (Tris), which might serve as RcdA effectors. Based on these structures, the ligand-binding pocket of RcdA was characterized in detail. The conservation of the amino acid residues forming the ligand-binding cavity was analyzed and the comprehensive search for RcdA structural homologs was performed. This analysis indicated that RcdA is structurally similar to multidrug-binding TetR family members, however, its ligand-binding cavity differs significantly from the pockets of its structural homologs. The interaction of RcdA with TMAO and Tris indicates that the protein might be involved in alkaline stress response.
- Institute of Molecular and Industrial Biotechnology, Faculty of Biotechnology and Food Sciences, Lodz University of Technology, Lodz, Poland.
Organizational Affiliation: