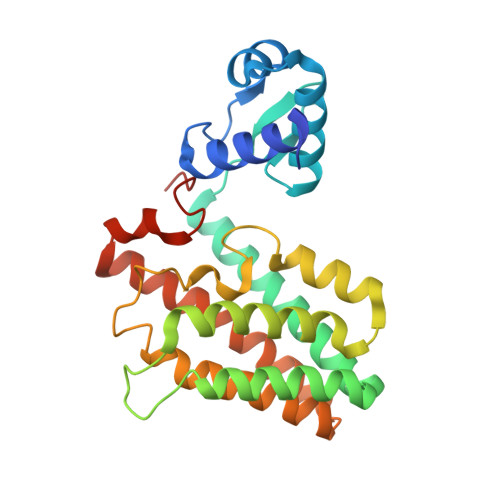
Characterization of the first tetrameric transcription factor of the GntR superfamily with allosteric regulation from the bacterial pathogen Agrobacterium fabrum.
Vigouroux, A., Meyer, T., Naretto, A., Legrand, P., Aumont-Nicaise, M., Di Cicco, A., Renoud, S., Dore, J., Levy, D., Vial, L., Lavire, C., Morera, S.(2021) Nucleic Acids Res 49: 529-546
- PubMed: 33313837
- DOI: https://doi.org/10.1093/nar/gkaa1181
- Primary Citation of Related Structures:
6Z74, 6ZA0, 6ZA3, 6ZA7, 6ZAB - PubMed Abstract:
A species-specific region, denoted SpG8-1b allowing hydroxycinnamic acids (HCAs) degradation is important for the transition between the two lifestyles (rhizospheric versus pathogenic) of the plant pathogen Agrobacterium fabrum. Indeed, HCAs can be either used as trophic resources and/or as induced-virulence molecules. The SpG8-1b region is regulated by two transcriptional regulators, namely, HcaR (Atu1422) and Atu1419. In contrast to HcaR, Atu1419 remains so far uncharacterized. The high-resolution crystal structures of two fortuitous citrate complexes, two DNA complexes and the apoform revealed that the tetrameric Atu1419 transcriptional regulator belongs to the VanR group of Pfam PF07729 subfamily of the large GntR superfamily. Until now, GntR regulators were described as dimers. Here, we showed that Atu1419 represses three genes of the HCAs catabolic pathway. We characterized both the effector and DNA binding sites and identified key nucleotides in the target palindrome. From promoter activity measurement using defective gene mutants, structural analysis and gel-shift assays, we propose N5,N10-methylenetetrahydrofolate as the effector molecule, which is not a direct product/substrate of the HCA degradation pathway. The Zn2+ ion present in the effector domain has both a structural and regulatory role. Overall, our work shed light on the allosteric mechanism of transcription employed by this GntR repressor.
- Université Paris-Saclay, CEA, CNRS, Institute for Integrative Biology of the Cell (I2BC), 91198 Gif-sur-Yvette, France.
Organizational Affiliation: