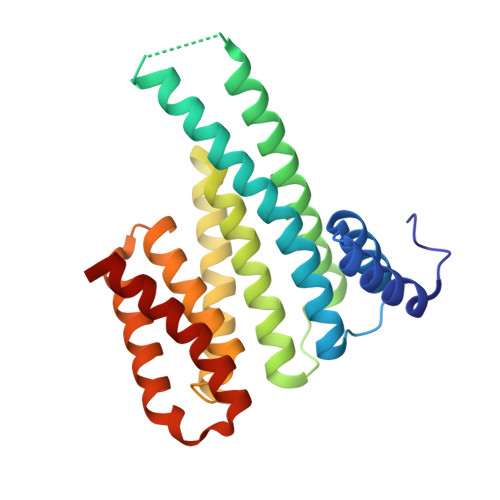
Understanding the interaction of 14-3-3 proteins with hDMX and hDM2: a structural and biophysical study.
Srdanovic, S., Wolter, M., Trinh, C.H., Ottmann, C., Warriner, S.L., Wilson, A.J.(2022) FEBS J
- PubMed: 35286747
- DOI: https://doi.org/10.1111/febs.16433
- Primary Citation of Related Structures:
6YR5, 6YR6, 6YR7 - PubMed Abstract:
p53 plays a critical role in regulating diverse biological processes: DNA repair, cell cycle arrest, apoptosis and senescence. The p53 pathway has therefore served as the focus of multiple drug-discovery efforts. p53 is negatively regulated by hDMX and hDM2; prior studies have identified 14-3-3 proteins as hDMX and hDM2 client proteins. 14-3-3 proteins are adaptor proteins that modulate localization, degradation and interactions of their targets in response to phosphorylation. Thus, 14-3-3 proteins may indirectly modulate the interaction between hDMX or hDM2 and p53 and represent potential targets for modulation of the p53 pathway. In this manuscript, we report on the biophysical and structural characterization of peptide/protein interactions that are representative of the interaction between 14-3-3 and hDMX or hDM2. The data establish that proximal phosphosites spaced ~20-25 residues apart in both hDMX and hDM2 co-operate to facilitate high-affinity 14-3-3 binding and provide structural insight that can be utilized in future stabilizer/inhibitor discovery efforts.
- Astbury Centre for Structural Molecular Biology, University of Leeds, UK.
Organizational Affiliation: