Novel Genetically Encoded Bright Positive Calcium Indicator NCaMP7 Based on the mNeonGreen Fluorescent Protein.
Subach, O.M., Sotskov, V.P., Plusnin, V.V., Gruzdeva, A.M., Barykina, N.V., Ivashkina, O.I., Anokhin, K.V., Nikolaeva, A.Y., Korzhenevskiy, D.A., Vlaskina, A.V., Lazarenko, V.A., Boyko, K.M., Rakitina, T.V., Varizhuk, A.M., Pozmogova, G.E., Podgorny, O.V., Piatkevich, K.D., Boyden, E.S., Subach, F.V.(2020) Int J Mol Sci 21
- PubMed: 32121243
- DOI: https://doi.org/10.3390/ijms21051644
- Primary Citation of Related Structures:
6XW2 - PubMed Abstract:
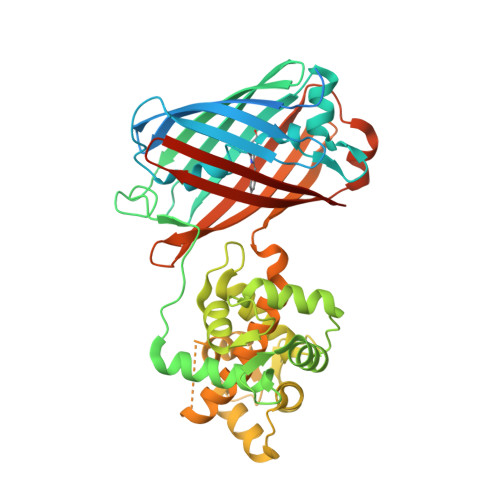
Green fluorescent genetically encoded calcium indicators (GECIs) are the most popular tool for visualization of calcium dynamics in vivo. However, most of them are based on the EGFP protein and have similar molecular brightnesses. The NTnC indicator, which is composed of the mNeonGreen fluorescent protein with the insertion of troponin C, has higher brightness as compared to EGFP-based GECIs, but shows a limited inverted response with an ΔF/F of 1. By insertion of a calmodulin/M13-peptide pair into the mNeonGreen protein, we developed a green GECI called NCaMP7. In vitro, NCaMP7 showed positive response with an ΔF/F of 27 and high affinity (K d of 125 nM) to calcium ions. NCaMP7 demonstrated a 1.7-fold higher brightness and similar calcium-association/dissociation dynamics compared to the standard GCaMP6s GECI in vitro. According to fluorescence recovery after photobleaching (FRAP) experiments, the NCaMP7 design partially prevented interactions of NCaMP7 with the intracellular environment. The NCaMP7 crystal structure was obtained at 1.75 Å resolution to uncover the molecular basis of its calcium ions sensitivity. The NCaMP7 indicator retained a high and fast response when expressed in cultured HeLa and neuronal cells. Finally, we successfully utilized the NCaMP7 indicator for in vivo visualization of grating-evoked and place-dependent neuronal activity in the visual cortex and the hippocampus of mice using a two-photon microscope and an NVista miniscope, respectively.
- National Research Center "Kurchatov Institute", Moscow 123182, Russia.
Organizational Affiliation: