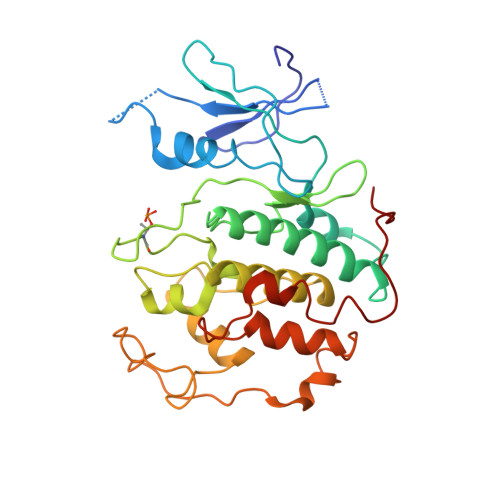
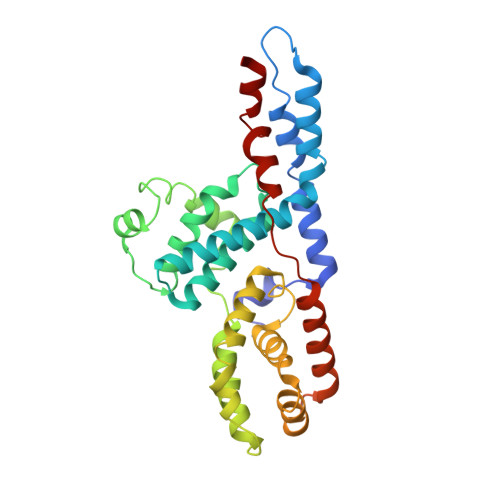
Structure of TFIIK for phosphorylation of CTD of RNA polymerase II.
van Eeuwen, T., Li, T., Kim, H.J., Gorbea Colon, J.J., Parker, M.I., Dunbrack, R.L., Garcia, B.A., Tsai, K.L., Murakami, K.(2021) Sci Adv 7
- PubMed: 33827808
- DOI: https://doi.org/10.1126/sciadv.abd4420
- Primary Citation of Related Structures:
6XI8, 7KUE - PubMed Abstract:
During transcription initiation, the general transcription factor TFIIH marks RNA polymerase II by phosphorylating Ser5 of the carboxyl-terminal domain (CTD) of Rpb1, which is followed by extensive modifications coupled to transcription elongation, mRNA processing, and histone dynamics. We have determined a 3.5-Å resolution cryo-electron microscopy (cryo-EM) structure of the TFIIH kinase module (TFIIK in yeast), which is composed of Kin28, Ccl1, and Tfb3, yeast homologs of CDK7, cyclin H, and MAT1, respectively. The carboxyl-terminal region of Tfb3 was lying at the edge of catalytic cleft of Kin28, where a conserved Tfb3 helix served to stabilize the activation loop in its active conformation. By combining the structure of TFIIK with the previous cryo-EM structure of the preinitiation complex, we extend the previously proposed model of the CTD path to the active site of TFIIK.
- Department of Biochemistry and Biophysics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Organizational Affiliation: