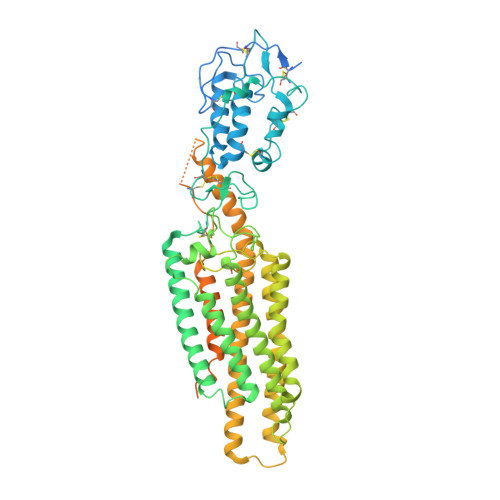
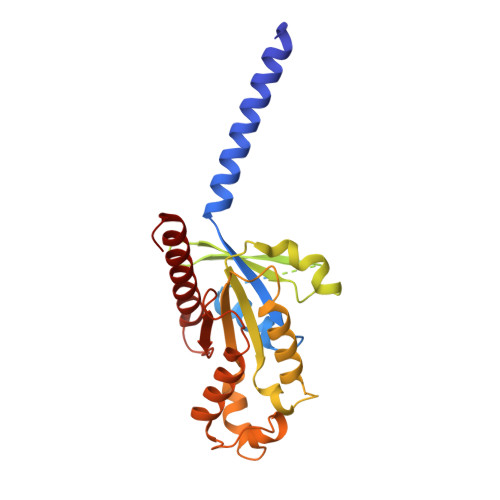
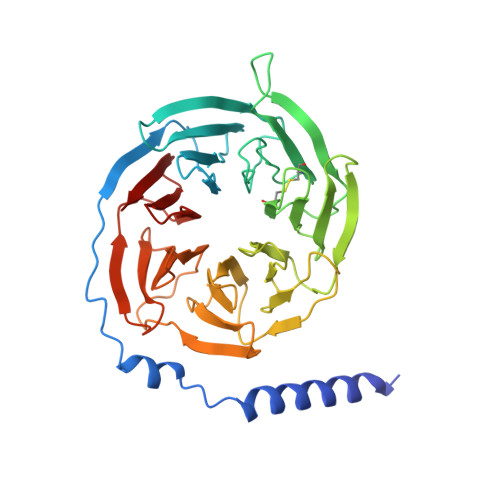
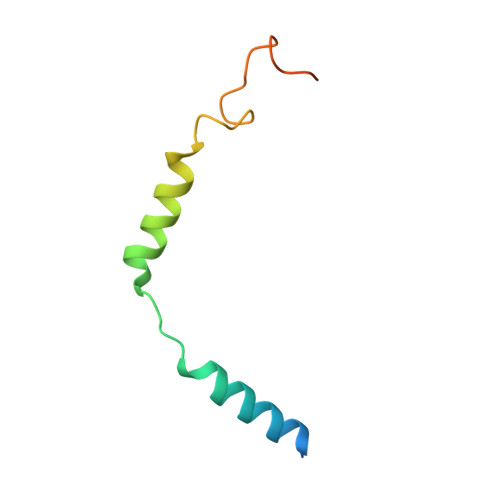
Sterols in an intramolecular channel of Smoothened mediate Hedgehog signaling.
Qi, X., Friedberg, L., De Bose-Boyd, R., Long, T., Li, X.(2020) Nat Chem Biol 16: 1368-1375
- PubMed: 32929279
- DOI: https://doi.org/10.1038/s41589-020-0646-2
- Primary Citation of Related Structures:
6XBJ, 6XBK, 6XBL, 6XBM - PubMed Abstract:
Smoothened (SMO), a class Frizzled G protein-coupled receptor (class F GPCR), transduces the Hedgehog signal across the cell membrane. Sterols can bind to its extracellular cysteine-rich domain (CRD) and to several sites in the seven transmembrane helices (7-TMs) of SMO. However, the mechanism by which sterols regulate SMO via multiple sites is unknown. Here we determined the structures of SMO-G i complexes bound to the synthetic SMO agonist (SAG) and to 24(S),25-epoxycholesterol (24(S),25-EC). A novel sterol-binding site in the extracellular extension of TM6 was revealed to connect other sites in 7-TMs and CRD, forming an intramolecular sterol channel from the middle side of 7-TMs to CRD. Additional structures of two gain-of-function variants, SMO D384R and SMO G111C/I496C , showed that blocking the channel at its midpoints allows sterols to occupy the binding sites in 7-TMs, thereby activating SMO. These data indicate that sterol transport through the core of SMO is a major regulator of SMO-mediated signaling.
- Department of Molecular Genetics, University of Texas Southwestern Medical Center, Dallas, TX, USA.
Organizational Affiliation: