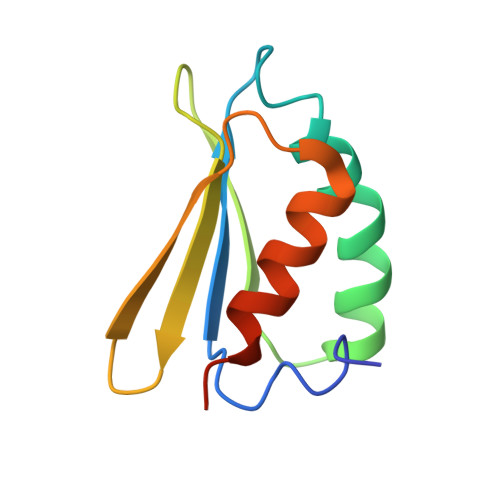
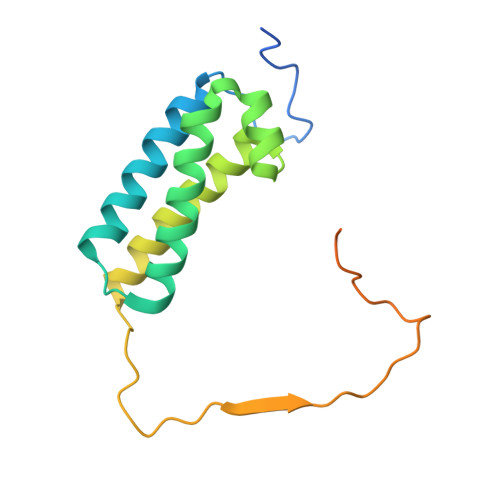
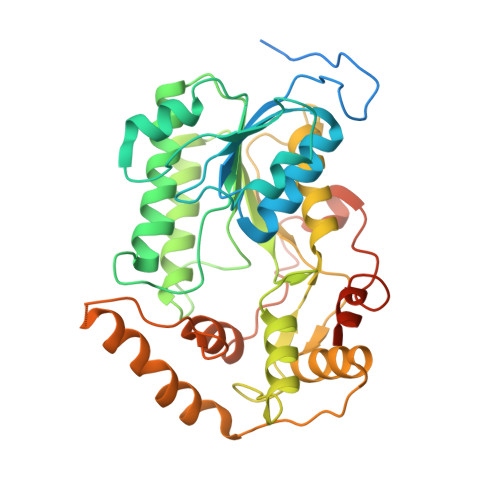
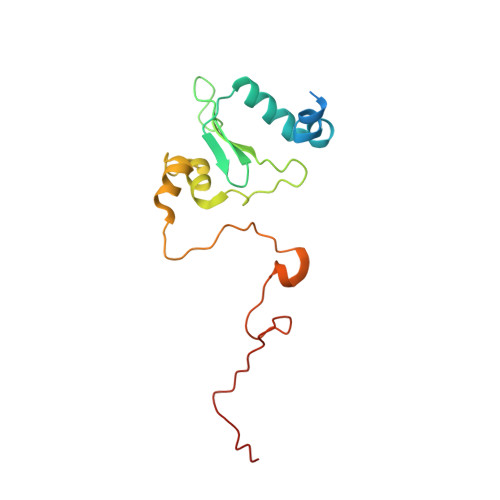
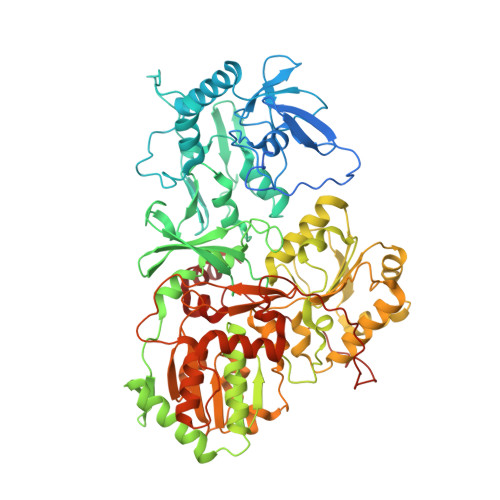
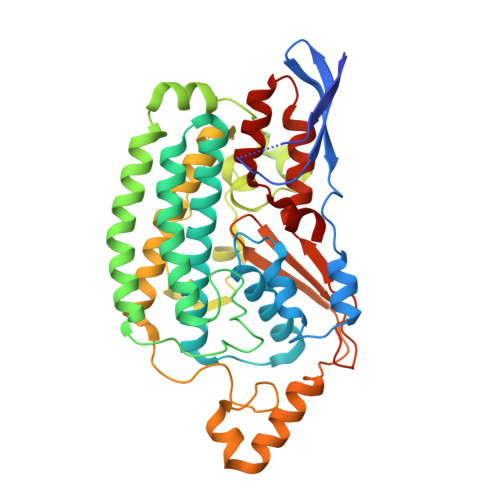
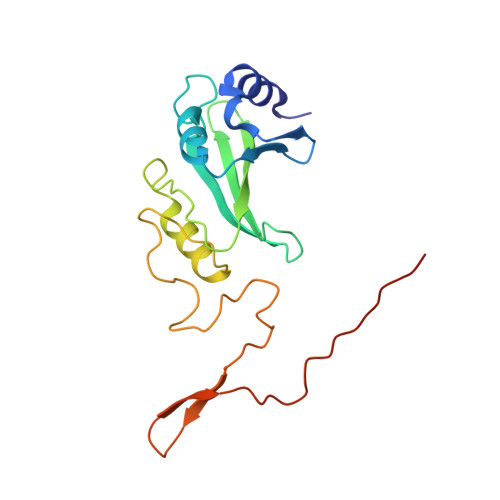
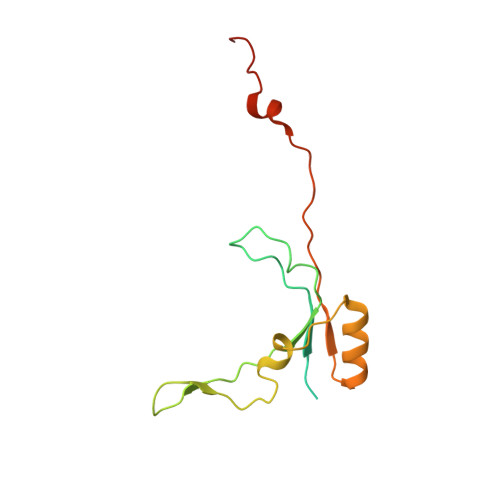
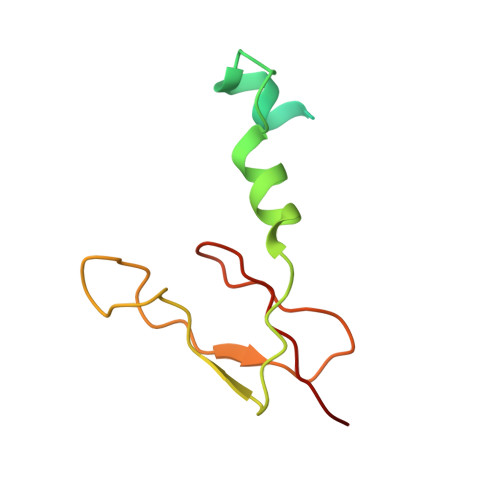
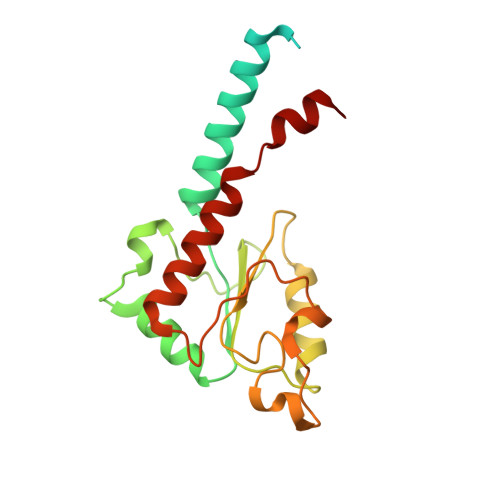
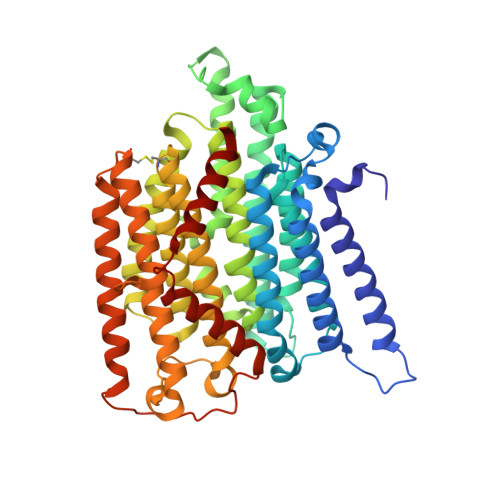
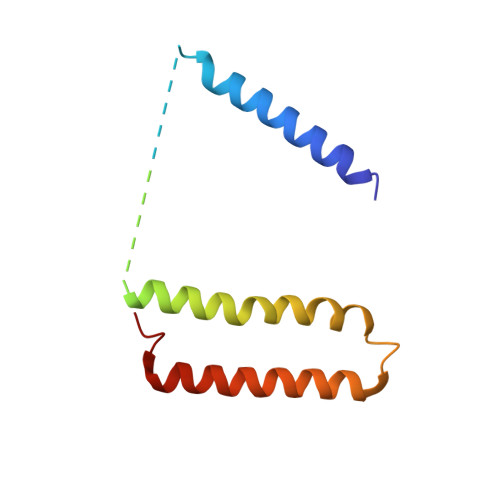
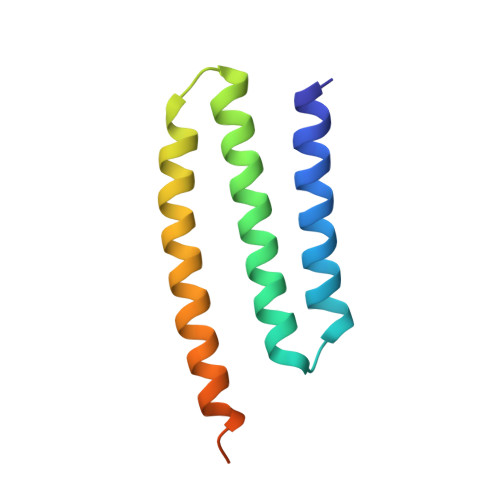
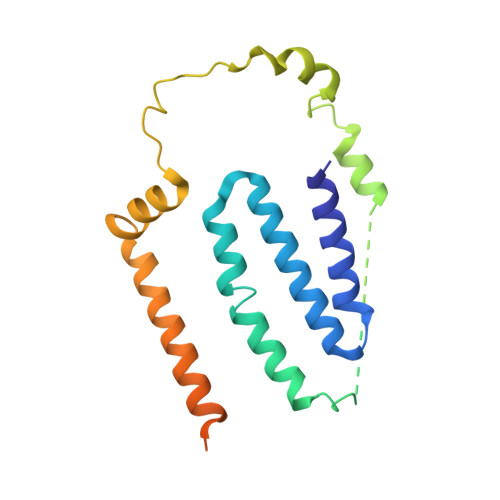
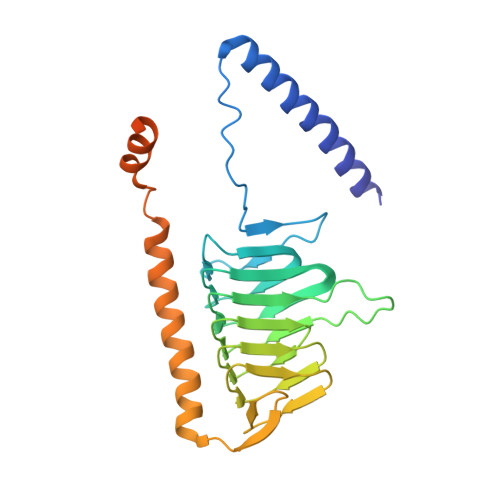
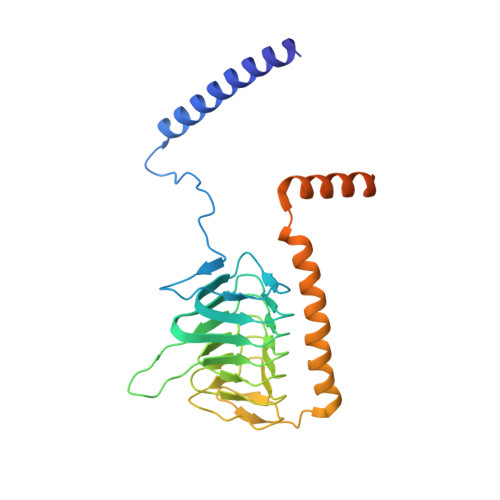
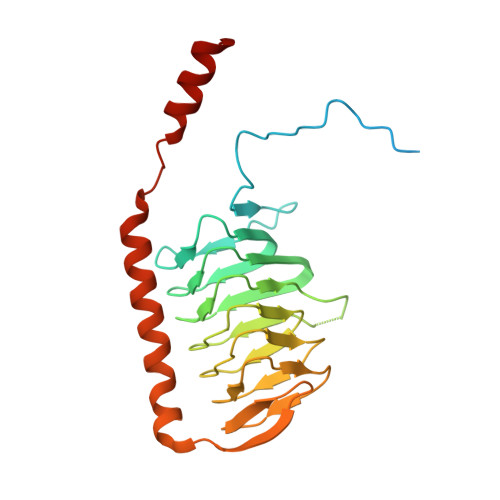
Atomic structure of a mitochondrial complex I intermediate from vascular plants.
Maldonado, M., Padavannil, A., Zhou, L., Guo, F., Letts, J.A.(2020) Elife 9
- PubMed: 32840211
- DOI: https://doi.org/10.7554/eLife.56664
- Primary Citation of Related Structures:
6X89 - PubMed Abstract:
Respiration, an essential metabolic process, provides cells with chemical energy. In eukaryotes, respiration occurs via the mitochondrial electron transport chain (mETC) composed of several large membrane-protein complexes. Complex I (CI) is the main entry point for electrons into the mETC. For plants, limited availability of mitochondrial material has curbed detailed biochemical and structural studies of their mETC. Here, we present the cryoEM structure of the known CI assembly intermediate CI* from Vigna radiata at 3.9 Å resolution. CI* contains CI's NADH-binding and CoQ-binding modules, the proximal-pumping module and the plant-specific γ-carbonic-anhydrase domain (γCA). Our structure reveals significant differences in core and accessory subunits of the plant complex compared to yeast, mammals and bacteria, as well as the details of the γCA domain subunit composition and membrane anchoring. The structure sheds light on differences in CI assembly across lineages and suggests potential physiological roles for CI* beyond assembly.
- Department of Molecular and Cellular Biology, University of California Davis, Davis, United States.
Organizational Affiliation: