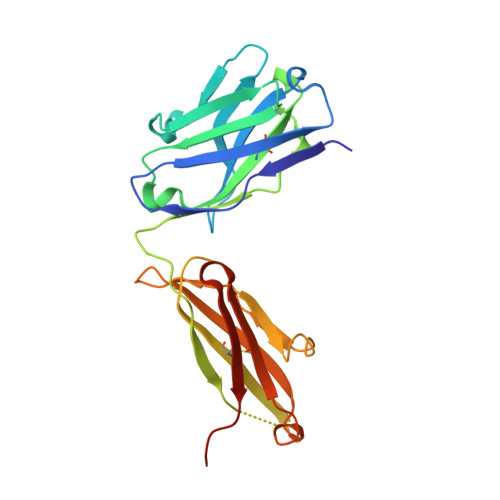
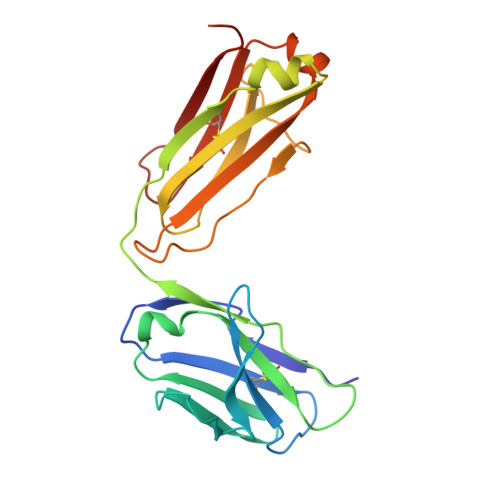
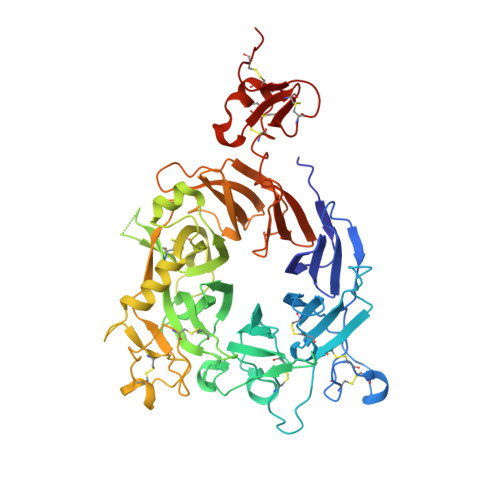
Discovery of amivantamab (JNJ-61186372), a bispecific antibody targeting EGFR and MET.
Neijssen, J., Cardoso, R.M.F., Chevalier, K.M., Wiegman, L., Valerius, T., Anderson, G.M., Moores, S.L., Schuurman, J., Parren, P.W.H.I., Strohl, W.R., Chiu, M.L.(2021) J Biological Chem 296: 100641-100641
- PubMed: 33839159
- DOI: https://doi.org/10.1016/j.jbc.2021.100641
- Primary Citation of Related Structures:
6WVZ - PubMed Abstract:
A bispecific antibody (BsAb) targeting the epidermal growth factor receptor (EGFR) and mesenchymal-epithelial transition factor (MET) pathways represents a novel approach to overcome resistance to targeted therapies in patients with non-small cell lung cancer. In this study, we sequentially screened a panel of BsAbs in a combinatorial approach to select the optimal bispecific molecule. The BsAbs were derived from different EGFR and MET parental monoclonal antibodies. Initially, molecules were screened for EGFR and MET binding on tumor cell lines and lack of agonistic activity toward MET. Hits were identified and further screened based on their potential to induce untoward cell proliferation and cross-phosphorylation of EGFR by MET via receptor colocalization in the absence of ligand. After the final step, we selected the EGFR and MET arms for the lead BsAb and added low fucose Fc engineering to generate amivantamab (JNJ-61186372). The crystal structure of the anti-MET Fab of amivantamab bound to MET was solved, and the interaction between the two molecules in atomic details was elucidated. Amivantamab antagonized the hepatocyte growth factor (HGF)-induced signaling by binding to MET Sema domain and thereby blocking HGF β-chain-Sema engagement. The amivantamab EGFR epitope was mapped to EGFR domain III and residues K443, K465, I467, and S468. Furthermore, amivantamab showed superior antitumor activity over small molecule EGFR and MET inhibitors in the HCC827-HGF in vivo model. Based on its unique mode of action, amivantamab may provide benefit to patients with malignancies associated with aberrant EGFR and MET signaling.
- Genmab, Utrecht, The Netherlands.
Organizational Affiliation: