Structural insights into differences in G protein activation by family A and family B GPCRs.
Hilger, D., Kumar, K.K., Hu, H., Pedersen, M.F., O'Brien, E.S., Giehm, L., Jennings, C., Eskici, G., Inoue, A., Lerch, M., Mathiesen, J.M., Skiniotis, G., Kobilka, B.K.(2020) Science 369
- PubMed: 32732395
- DOI: https://doi.org/10.1126/science.aba3373
- Primary Citation of Related Structures:
6WPW - PubMed Abstract:
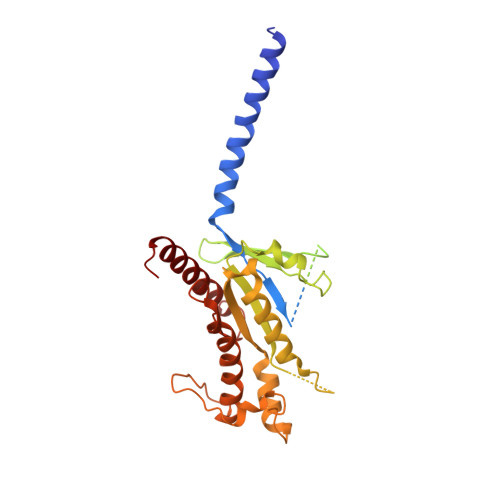
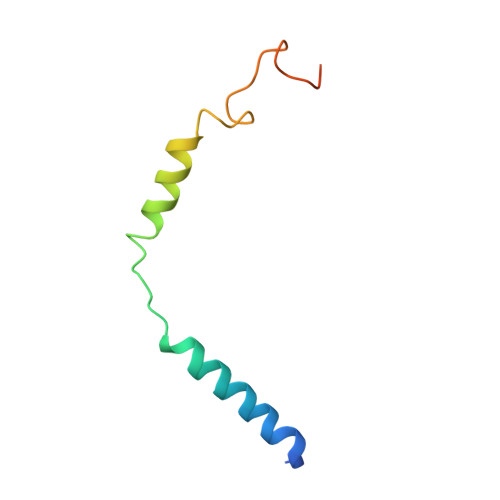
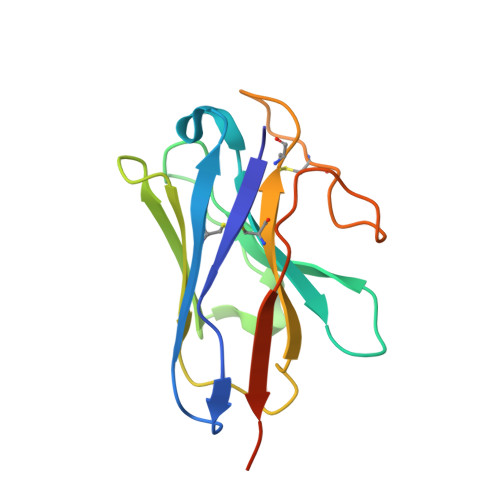
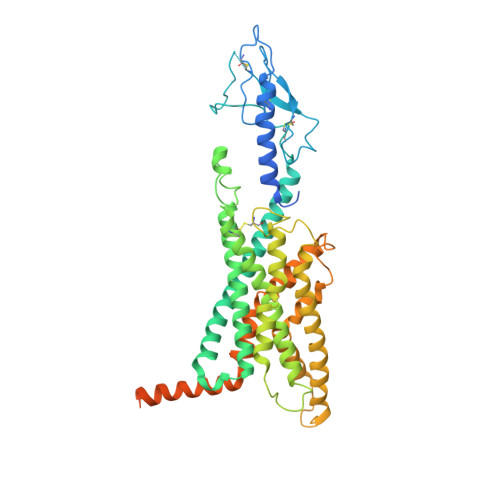
Family B heterotrimeric guanine nucleotide-binding protein (G protein)-coupled receptors (GPCRs) play important roles in carbohydrate metabolism. Recent structures of family B GPCR-G s protein complexes reveal a disruption in the α-helix of transmembrane segment 6 (TM6) not observed in family A GPCRs. To investigate the functional impact of this structural difference, we compared the structure and function of the glucagon receptor (GCGR; family B) with the β 2 adrenergic receptor (β 2 AR; family A). We determined the structure of the GCGR-G s complex by means of cryo-electron microscopy at 3.1-angstrom resolution. This structure shows the distinct break in TM6. Guanosine triphosphate (GTP) turnover, guanosine diphosphate release, GTP binding, and G protein dissociation studies revealed much slower rates for G protein activation by the GCGR compared with the β 2 AR. Fluorescence and double electron-electron resonance studies suggest that this difference is due to the inability of agonist alone to induce a detectable outward movement of the cytoplasmic end of TM6.
- Department of Molecular and Cellular Physiology, Stanford University School of Medicine, 279 Campus Drive, Stanford, CA 94305, USA.
Organizational Affiliation: