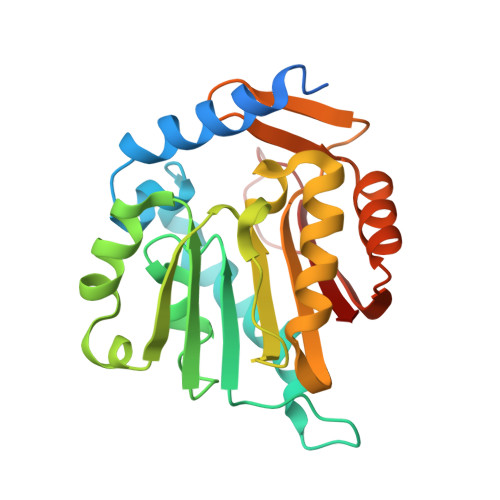
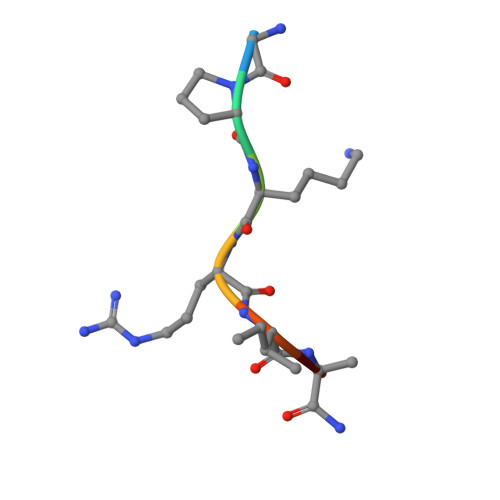
Probing the Plasticity in the Active Site of Protein N-terminal Methyltransferase 1 Using Bisubstrate Analogues.
Chen, D., Dong, C., Dong, G., Srinivasan, K., Min, J., Noinaj, N., Huang, R.(2020) J Med Chem 63: 8419-8431
- PubMed: 32605369
- DOI: https://doi.org/10.1021/acs.jmedchem.0c00770
- Primary Citation of Related Structures:
6PVB, 6WJ7 - PubMed Abstract:
The bisubstrate analogue strategy is a promising approach to develop potent and selective inhibitors for protein methyltransferases. Herein, the interactions of a series of bisubstrate analogues with protein N-terminal methyltransferase 1 (NTMT1) were examined to probe the molecular properties of the active site of NTMT1. Our results indicate that a 2-C to 4-C atom linker enables its respective bisubstrate analogue to occupy both substrate- and cofactor-binding sites of NTMT1, but the bisubstrate analogue with a 5-C atom linker only interacts with the substrate-binding site and functions as a substrate. Furthermore, the 4-C atom linker is the optimal and produces the most potent inhibitor ( K i,app = 130 ± 40 pM) for NTMT1 to date, displaying more than 3000-fold selectivity for other methyltransferases and even for its homologue NTMT2. This study reveals the molecular basis for the plasticity of the active site of NTMT1. Additionally, our study outlines general guidance on the development of bisubstrate inhibitors for any methyltransferases.
- Department of Medicinal Chemistry and Molecular Pharmacology, Center for Cancer Research, Institute for Drug Discovery, Purdue University, West Lafayette, Indiana 47907, United States.
Organizational Affiliation: