Topoisomerase Inhibitors Addressing Fluoroquinolone Resistance in Gram-Negative Bacteria.
Skepper, C.K., Armstrong, D., Balibar, C.J., Bauer, D., Bellamacina, C., Benton, B.M., Bussiere, D., De Pascale, G., De Vicente, J., Dean, C.R., Dhumale, B., Fisher, L.M., Fuller, J., Fulsunder, M., Holder, L.M., Hu, C., Kantariya, B., Lapointe, G., Leeds, J.A., Li, X., Lu, P., Lvov, A., Ma, S., Madhavan, S., Malekar, S., McKenney, D., Mergo, W., Metzger, L., Moser, H.E., Mutnick, D., Noeske, J., Osborne, C., Patel, A., Patel, D., Patel, T., Prajapati, K., Prosen, K.R., Reck, F., Richie, D.L., Rico, A., Sanderson, M.R., Satasia, S., Sawyer, W.S., Selvarajah, J., Shah, N., Shanghavi, K., Shu, W., Thompson, K.V., Traebert, M., Vala, A., Vala, L., Veselkov, D.A., Vo, J., Wang, M., Widya, M., Williams, S.L., Xu, Y., Yue, Q., Zang, R., Zhou, B., Rivkin, A.(2020) J Med Chem 63: 7773-7816
- PubMed: 32634310
- DOI: https://doi.org/10.1021/acs.jmedchem.0c00347
- Primary Citation of Related Structures:
6WAA - PubMed Abstract:
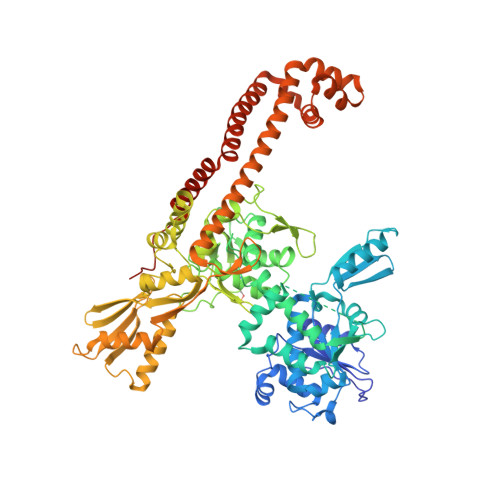
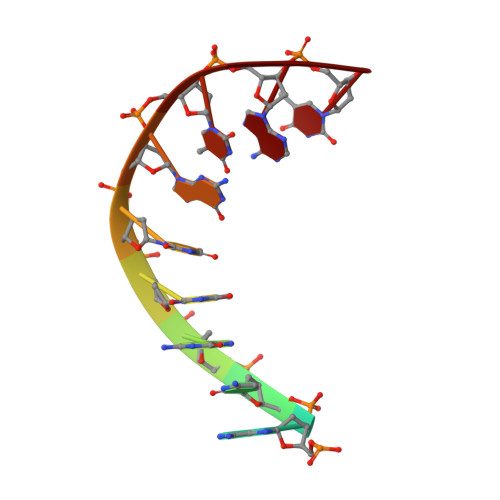
Since their discovery over 5 decades ago, quinolone antibiotics have found enormous success as broad spectrum agents that exert their activity through dual inhibition of bacterial DNA gyrase and topoisomerase IV. Increasing rates of resistance, driven largely by target-based mutations in the GyrA/ParC quinolone resistance determining region, have eroded the utility and threaten the future use of this vital class of antibiotics. Herein we describe the discovery and optimization of a series of 4-(aminomethyl)quinolin-2(1 H )-ones, exemplified by 34 , that inhibit bacterial DNA gyrase and topoisomerase IV and display potent activity against ciprofloxacin-resistant Gram-negative pathogens. X-ray crystallography reveals that 34 occupies the classical quinolone binding site in the topoisomerase IV-DNA cleavage complex but does not form significant contacts with residues in the quinolone resistance determining region.
- Novartis Institutes for BioMedical Research, Emeryville, California 94608, United States.
Organizational Affiliation: