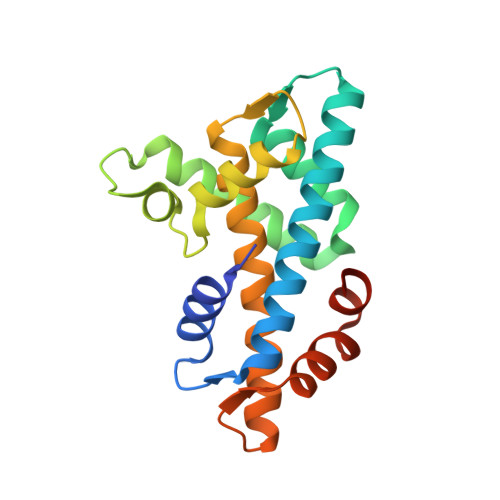
Structure of a single-chain H2A/H2B dimer.
Warren, C., Bonanno, J.B., Almo, S.C., Shechter, D.(2020) Acta Crystallogr F Struct Biol Commun 76: 194-198
- PubMed: 32356520
- DOI: https://doi.org/10.1107/S2053230X20004604
- Primary Citation of Related Structures:
6W4L - PubMed Abstract:
Chromatin is the complex assembly of nucleic acids and proteins that makes up the physiological form of the eukaryotic genome. The nucleosome is the fundamental repeating unit of chromatin, and is composed of ∼147 bp of DNA wrapped around a histone octamer formed by two copies of each core histone: H2A, H2B, H3 and H4. Prior to nucleosome assembly, and during histone eviction, histones are typically assembled into soluble H2A/H2B dimers and H3/H4 dimers and tetramers. A multitude of factors interact with soluble histone dimers and tetramers, including chaperones, importins, histone-modifying enzymes and chromatin-remodeling enzymes. It is still unclear how many of these proteins recognize soluble histones; therefore, there is a need for new structural tools to study non-nucleosomal histones. Here, a single-chain, tailless Xenopus H2A/H2B dimer was created by directly fusing the C-terminus of H2B to the N-terminus of H2A. It is shown that this construct (termed scH2BH2A) is readily expressed in bacteria and can be purified under non-denaturing conditions. A 1.31 Å resolution crystal structure of scH2BH2A shows that it adopts a conformation that is nearly identical to that of nucleosomal H2A/H2B. This new tool is likely to facilitate future structural studies of many H2A/H2B-interacting proteins.
- Department of Biochemistry, Albert Einstein College of Medicine, 1300 Morris Park Avenue, Bronx, NY 10461, USA.
Organizational Affiliation: