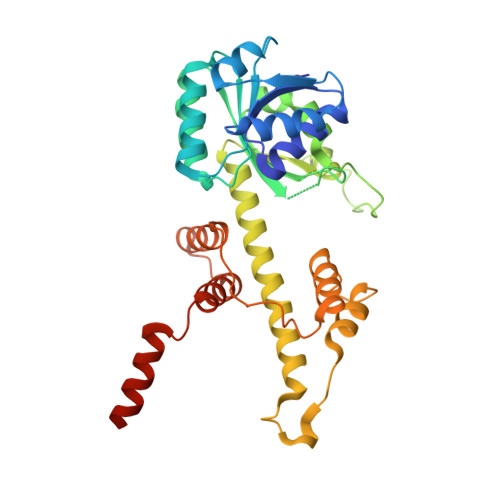
The structure of a novel membrane-associated 6-phosphogluconate dehydrogenase from Gluconacetobacter diazotrophicus (Gd6PGD) reveals a subfamily of short-chain 6PGDs.
Sarmiento-Pavia, P.D., Rodriguez-Hernandez, A., Rodriguez-Romero, A., Sosa-Torres, M.E.(2021) FEBS J 288: 1286-1304
- PubMed: 32621793
- DOI: https://doi.org/10.1111/febs.15472
- Primary Citation of Related Structures:
6VPB - PubMed Abstract:
The enzyme 6-phosphogluconate dehydrogenase catalyzes the conversion of 6-phosphogluconate to ribulose-5-phosphate. It represents an important reaction in the oxidative pentose phosphate pathway, producing a ribose precursor essential for nucleotide and nucleic acid synthesis. We succeeded, for the first time, to determine the three-dimensional structure of this enzyme from an acetic acid bacterium, Gluconacetobacter diazotrophicus (Gd6PGD). Active Gd6PGD, a homodimer (70 kDa), was present in both the soluble and the membrane fractions of the nitrogen-fixing microorganism. The Gd6PGD belongs to the newly described subfamily of short-chain (333 AA) 6PGDs, compared to the long-chain subfamily (480 AA; e.g., Ovis aries, Homo sapiens). The shorter amino acid sequence in Gd6PGD induces the exposition of hydrophobic residues in the C-terminal domain. This distinct structural feature is key for the protein to associate with the membrane. Furthermore, in terms of function, the short-chain 6PGD seems to prefer NAD + over NADP + , delivering NADH to the membrane-bound NADH dehydrogenase of the microorganisms required by the terminal oxidases to reduce dioxygen to water for energy conservation. ENZYME: ECnonbreakingspace1.1.1.343. DATABASE: Structural data are available in PDB database under the accession number 6VPB.
- Facultad de Química, Universidad Nacional Autónoma de México, Coyoacán, Mexico.
Organizational Affiliation: