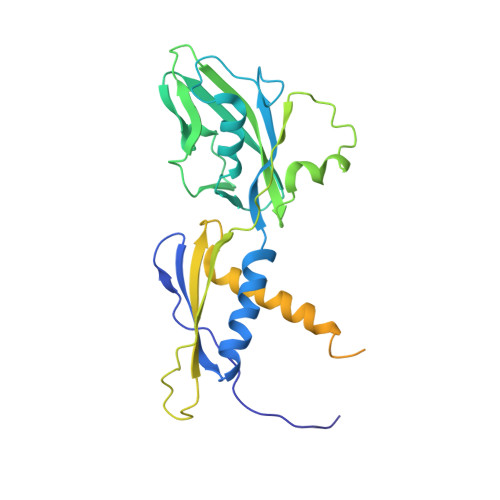
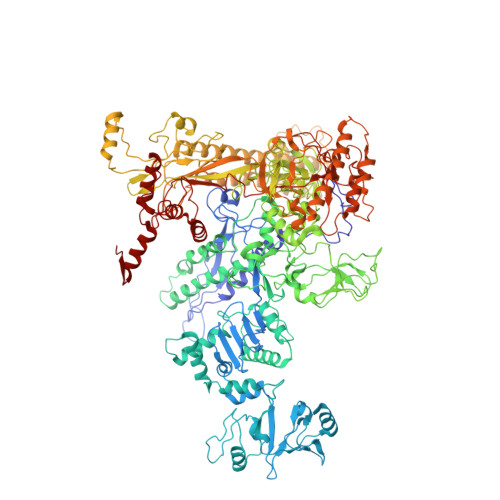
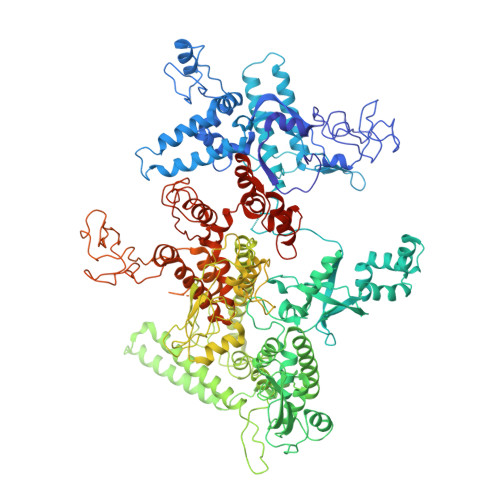
Evaluation of Bacterial RNA Polymerase Inhibitors in a Staphylococcus aureus -Based Wound Infection Model in SKH1 Mice.
Haupenthal, J., Kautz, Y., Elgaher, W.A.M., Patzold, L., Rohrig, T., Laschke, M.W., Tschernig, T., Hirsch, A.K.H., Molodtsov, V., Murakami, K.S., Hartmann, R.W., Bischoff, M.(2020) ACS Infect Dis 6: 2573-2581
- PubMed: 32886885
- DOI: https://doi.org/10.1021/acsinfecdis.0c00034
- Primary Citation of Related Structures:
6VJS - PubMed Abstract:
Chronic wounds infected with pathogens such as Staphylococcus aureus represent a worldwide health concern, especially in patients with a compromised immune system. As antimicrobial resistance has become an immense global problem, novel antibiotics are urgently needed. One strategy to overcome this threatening situation is the search for drugs targeting novel binding sites on essential and validated enzymes such as the bacterial RNA polymerase (RNAP). In this work, we describe the establishment of an in vivo wound infection model based on the pathogen S. aureus and hairless Crl:SKH1-Hrhr (SKH1) mice. The model proved to be a valuable preclinical tool to study selected RNAP inhibitors after topical application. While rifampicin showed a reduction in the loss of body weight induced by the bacteria, an acceleration of wound healing kinetics, and a reduced number of colony forming units in the wound, the ureidothiophene-2-carboxylic acid 1 was inactive under in vivo conditions, probably due to strong plasma protein binding. The cocrystal structure of compound 1 with RNAP, that we hereby also present, will be of great value for applying appropriate structural modifications to further optimize the compound, especially in terms of plasma protein binding.
- Department of Drug Design and Optimization, Helmholtz Institute for Pharmaceutical Research Saarland (HIPS)-Helmholtz Centre for Infection Research (HZI), Campus Building E8.1, 66123 Saarbrücken, Saarland, Germany.
Organizational Affiliation: