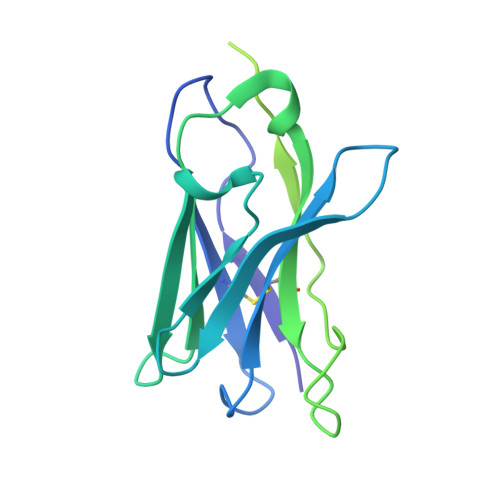
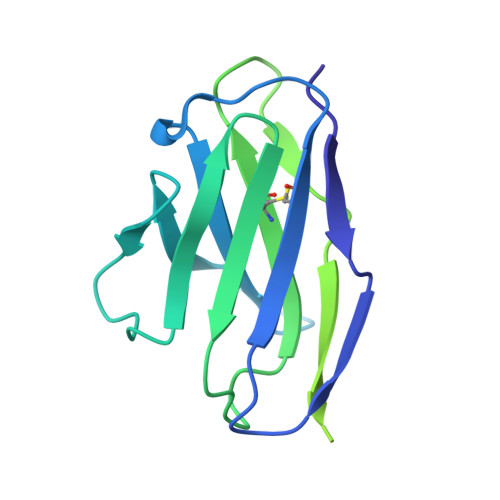
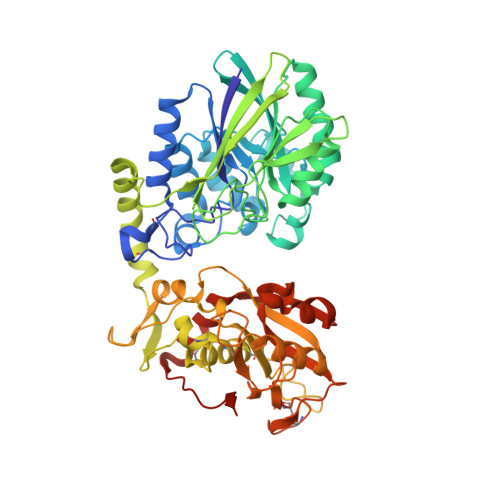
A highly potent CD73 biparatopic antibody blocks organization of the enzyme active site through dual mechanisms.
Stefano, J.E., Lord, D.M., Zhou, Y., Jaworski, J., Hopke, J., Travaline, T., Zhang, N., Wong, K., Lennon, A., He, T., Bric-Furlong, E., Cherrie, C., Magnay, T., Remy, E., Brondyk, W., Qiu, H., Radosevic, K.(2020) J Biological Chem 295: 18379-18389
- PubMed: 33122192
- DOI: https://doi.org/10.1074/jbc.RA120.012395
- Primary Citation of Related Structures:
6VC9, 6VCA - PubMed Abstract:
The dimeric ectonucleotidase CD73 catalyzes the hydrolysis of AMP at the cell surface to form adenosine, a potent suppressor of the immune response. Blocking CD73 activity in the tumor microenvironment can have a beneficial effect on tumor eradication and is a promising approach for cancer therapy. Biparatopic antibodies binding different regions of CD73 may be a means to antagonize its enzymatic activity. A panel of biparatopic antibodies representing the pairwise combination of 11 parental monoclonal antibodies against CD73 was generated by Fab-arm exchange. Nine variants vastly exceeded the potency of their parental antibodies with ≥90% inhibition of activity and subnanomolar EC 50 values. Pairing the Fabs of parents with nonoverlapping epitopes was both sufficient and necessary whereas monovalent antibodies were poor inhibitors. Some parental antibodies yielded potent biparatopics with multiple partners, one of which (TB19) producing the most potent. The structure of the TB19 Fab with CD73 reveals that it blocks alignment of the N- and C-terminal CD73 domains necessary for catalysis. A separate structure of CD73 with a Fab (TB38) which complements TB19 in a particularly potent biparatopic shows its binding to a nonoverlapping site on the CD73 N-terminal domain. Structural modeling demonstrates a TB19/TB38 biparatopic antibody would be unable to bind the CD73 dimer in a bivalent manner, implicating crosslinking of separate CD73 dimers in its mechanism of action. This ability of a biparatopic antibody to both crosslink CD73 dimers and fix them in an inactive conformation thus represents a highly effective mechanism for the inhibition of CD73 activity.
- Biologics Research, Sanofi R&D Framingham, Massachusetts, USA.
Organizational Affiliation: