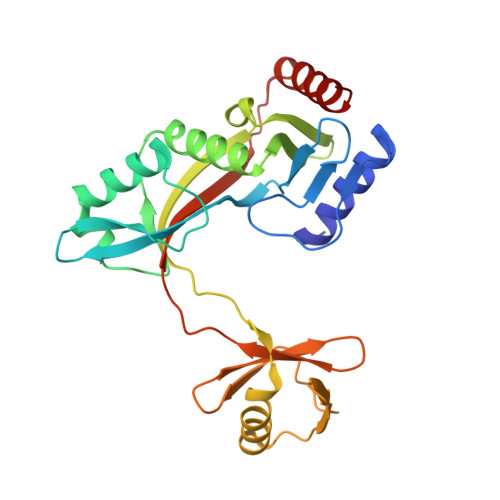
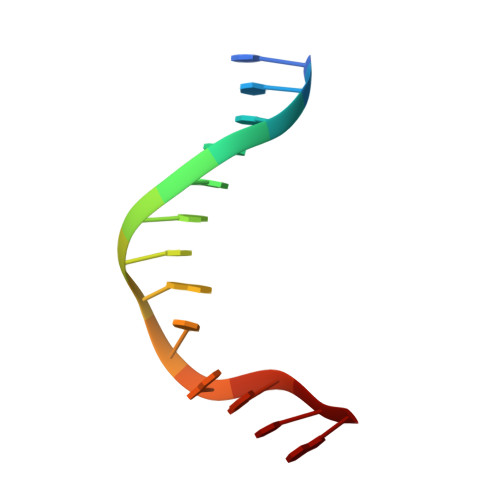
Structure of HhaI endonuclease with cognate DNA at an atomic resolution of 1.0 angstrom.
Horton, J.R., Yang, J., Zhang, X., Petronzio, T., Fomenkov, A., Wilson, G.G., Roberts, R.J., Cheng, X.(2020) Nucleic Acids Res 48: 1466-1478
- PubMed: 31879785
- DOI: https://doi.org/10.1093/nar/gkz1195
- Primary Citation of Related Structures:
6UKE, 6UKF, 6UKG, 6UKH, 6UKI - PubMed Abstract:
HhaI, a Type II restriction endonuclease, recognizes the symmetric sequence 5'-GCG↓C-3' in duplex DNA and cleaves ('↓') to produce fragments with 2-base, 3'-overhangs. We determined the structure of HhaI in complex with cognate DNA at an ultra-high atomic resolution of 1.0 Å. Most restriction enzymes act as dimers with two catalytic sites, and cleave the two strands of duplex DNA simultaneously, in a single binding event. HhaI, in contrast, acts as a monomer with only one catalytic site, and cleaves the DNA strands sequentially, one after the other. HhaI comprises three domains, each consisting of a mixed five-stranded β sheet with a defined function. The first domain contains the catalytic-site; the second contains residues for sequence recognition; and the third contributes to non-specific DNA binding. The active-site belongs to the 'PD-D/EXK' superfamily of nucleases and contains the motif SD-X11-EAK. The first two domains are similar in structure to two other monomeric restriction enzymes, HinP1I (G↓CGC) and MspI (C↓CGG), which produce fragments with 5'-overhangs. The third domain, present only in HhaI, shifts the positions of the recognition residues relative to the catalytic site enabling this enzyme to cleave the recognition sequence at a different position. The structure of M.HhaI, the biological methyltransferase partner of HhaI, was determined earlier. Together, these two structures represent the first natural pair of restriction-modification enzymes to be characterized in atomic detail.
- Department of Epigenetics & Molecular Carcinogenesis, The University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Organizational Affiliation: