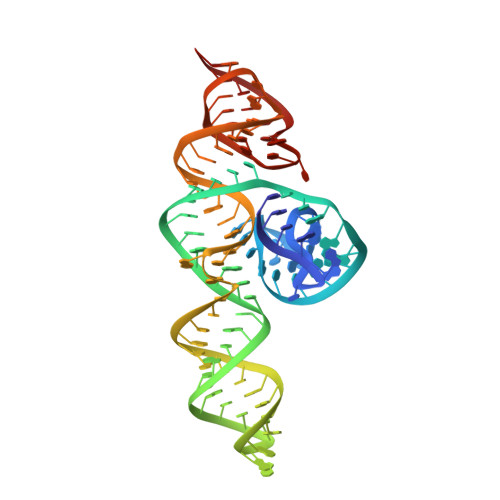
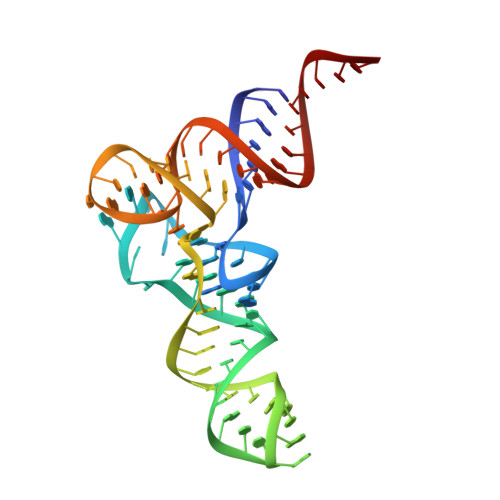High-affinity recognition of specific tRNAs by an mRNA anticodon-binding groove.
Suddala, K.C., Zhang, J.(2019) Nat Struct Mol Biol 26: 1114-1122
- PubMed: 31792448
- DOI: https://doi.org/10.1038/s41594-019-0335-6
- Primary Citation of Related Structures:
6UFM - PubMed Abstract:
T-box riboswitches are modular bacterial noncoding RNAs that sense and regulate amino acid availability through direct interactions with tRNAs. Between the 5' anticodon-binding stem I domain and the 3' amino acid sensing domains of most T-boxes lies the stem II domain of unknown structure and function. Here, we report a 2.8-Å cocrystal structure of the Nocardia farcinica ileS T-box in complex with its cognate tRNA Ile . The structure reveals a perpendicularly arranged ultrashort stem I containing a K-turn and an elongated stem II bearing an S-turn. Both stems rest against a compact pseudoknot, dock via an extended ribose zipper and jointly create a binding groove specific to the anticodon of its cognate tRNA. Contrary to proposed distal contacts to the tRNA elbow region, stem II locally reinforces the codon-anticodon interactions between stem I and tRNA, achieving low-nanomolar affinity. This study illustrates how mRNA junctions can create specific binding sites for interacting RNAs of prescribed sequence and structure.
- Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD, USA.
Organizational Affiliation: