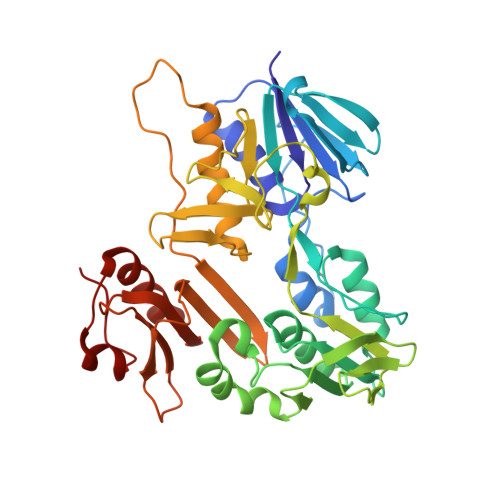
Expression, purification and crystal structure determination of a ferredoxin reductase from the actinobacterium Thermobifida fusca.
Rodriguez Buitrago, J.A., Klunemann, T., Blankenfeldt, W., Schallmey, A.(2020) Acta Crystallogr F Struct Biol Commun 76: 334-340
- PubMed: 32744244
- DOI: https://doi.org/10.1107/S2053230X2000922X
- Primary Citation of Related Structures:
6TUK - PubMed Abstract:
The ferredoxin reductase FdR9 from Thermobifida fusca, a member of the oxygenase-coupled NADH-dependent ferredoxin reductase (FNR) family, catalyses electron transfer from NADH to its physiological electron acceptor ferredoxin. It forms part of a putative three-component cytochrome P450 monooxygenase system in T. fusca comprising CYP222A1 and the [3Fe-4S]-cluster ferredoxin Fdx8 as well as FdR9. Here, FdR9 was overexpressed and purified and its crystal structure was determined at 1.9 Å resolution. The overall structure of FdR9 is similar to those of other members of the FNR family and is composed of an FAD-binding domain, an NAD-binding domain and a C-terminal domain. Activity measurements with FdR9 confirmed a strong preference for NADH as the cofactor. Comparison of the FAD- and NAD-binding domains of FdR9 with those of other ferredoxin reductases revealed the presence of conserved sequence motifs in the FAD-binding domain as well as several highly conserved residues involved in FAD and NAD cofactor binding. Moreover, the NAD-binding site of FdR9 contains a modified Rossmann-fold motif, GxSxxS, instead of the classical GxGxxG motif.
- Institute for Biochemistry, Biotechnology and Bioinformatics, Technical University Braunschweig, Spielmannstrasse 7, 38106 Braunschweig, Germany.
Organizational Affiliation: