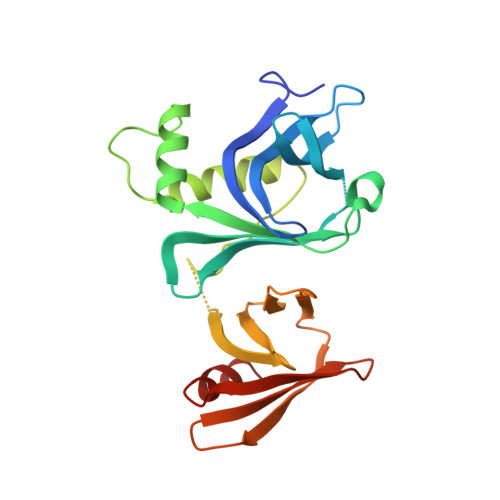
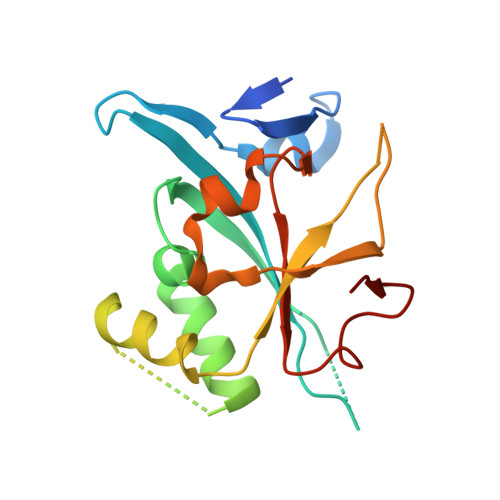
The box C/D snoRNP assembly factor Bcd1 interacts with the histone chaperone Rtt106 and controls its transcription dependent activity.
Bragantini, B., Charron, C., Bourguet, M., Paul, A., Tiotiu, D., Rothe, B., Marty, H., Terral, G., Hessmann, S., Decourty, L., Chagot, M.E., Strub, J.M., Massenet, S., Bertrand, E., Quinternet, M., Saveanu, C., Cianferani, S., Labialle, S., Manival, X., Charpentier, B.(2021) Nat Commun 12: 1859-1859
- PubMed: 33767140
- DOI: https://doi.org/10.1038/s41467-021-22077-4
- Primary Citation of Related Structures:
6NZ2, 6THL - PubMed Abstract:
Biogenesis of eukaryotic box C/D small nucleolar ribonucleoproteins initiates co-transcriptionally and requires the action of the assembly machinery including the Hsp90/R2TP complex, the Rsa1p:Hit1p heterodimer and the Bcd1 protein. We present genetic interactions between the Rsa1p-encoding gene and genes involved in chromatin organization including RTT106 that codes for the H3-H4 histone chaperone Rtt106p controlling H3K56ac deposition. We show that Bcd1p binds Rtt106p and controls its transcription-dependent recruitment by reducing its association with RNA polymerase II, modulating H3K56ac levels at gene body. We reveal the 3D structures of the free and Rtt106p-bound forms of Bcd1p using nuclear magnetic resonance and X-ray crystallography. The interaction is also studied by a combination of biophysical and proteomic techniques. Bcd1p interacts with a region that is distinct from the interaction interface between the histone chaperone and histone H3. Our results are evidence for a protein interaction interface for Rtt106p that controls its transcription-associated activity.
- Université de Lorraine, CNRS, IMoPA, Nancy, France.
Organizational Affiliation: