Tutuilamides A-C: Vinyl-Chloride-Containing Cyclodepsipeptides from Marine Cyanobacteria with Potent Elastase Inhibitory Properties.
Keller, L., Canuto, K.M., Liu, C., Suzuki, B.M., Almaliti, J., Sikandar, A., Naman, C.B., Glukhov, E., Luo, D., Duggan, B.M., Luesch, H., Koehnke, J., O'Donoghue, A.J., Gerwick, W.H.(2020) ACS Chem Biol 15: 751-757
- PubMed: 31935054
- DOI: https://doi.org/10.1021/acschembio.9b00992
- Primary Citation of Related Structures:
6TH7 - PubMed Abstract:
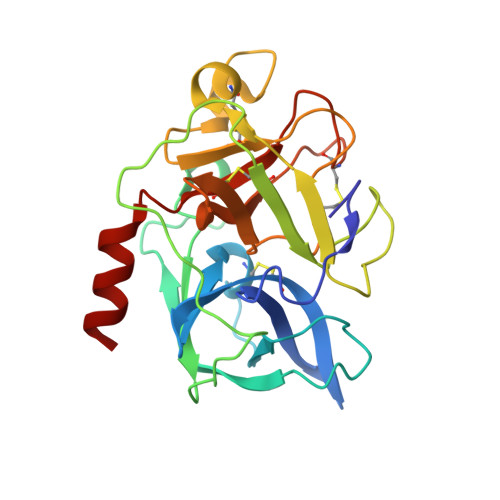
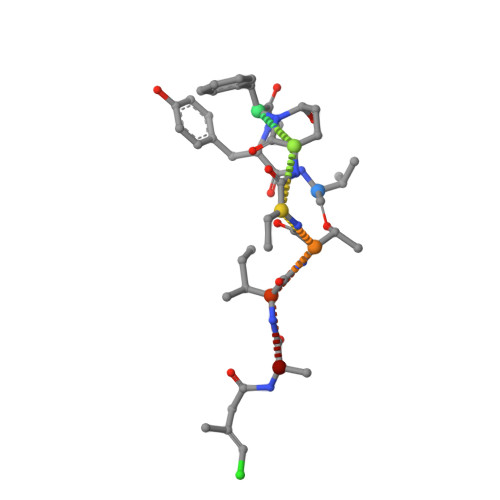
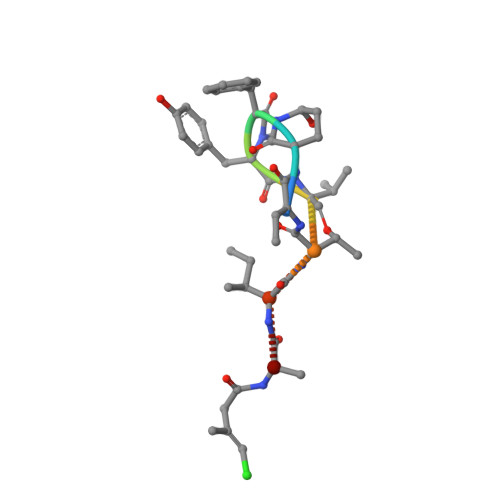
Marine cyanobacteria (blue-green algae) have been shown to possess an enormous capacity to produce structurally diverse natural products that exhibit a broad spectrum of potent biological activities, including cytotoxic, antifungal, antiparasitic, antiviral, and antibacterial activities. Using mass-spectrometry-guided fractionation together with molecular networking, cyanobacterial field collections from American Samoa and Palmyra Atoll yielded three new cyclic peptides, tutuilamides A-C. Their structures were established by spectroscopic techniques including 1D and 2D NMR, HR-MS, and chemical derivatization. Structure elucidation was facilitated by employing advanced NMR techniques including nonuniform sampling in combination with the 1,1-ADEQUATE experiment. These cyclic peptides are characterized by the presence of several unusual residues including 3-amino-6-hydroxy-2-piperidone and 2-amino-2-butenoic acid, together with a novel vinyl chloride-containing residue. Tutuilamides A-C show potent elastase inhibitory activity together with moderate potency in H-460 lung cancer cell cytotoxicity assays. The binding mode to elastase was analyzed by X-ray crystallography revealing a reversible binding mode similar to the natural product lyngbyastatin 7. The presence of an additional hydrogen bond with the amino acid backbone of the flexible side chain of tutuilamide A, compared to lyngbyastatin 7, facilitates its stabilization in the elastase binding pocket and possibly explains its enhanced inhibitory potency.
- Center for Marine Biotechnology and Biomedicine, Scripps Institution of Oceanography, University of California San Diego, La Jolla, California 92093, United States.
Organizational Affiliation: