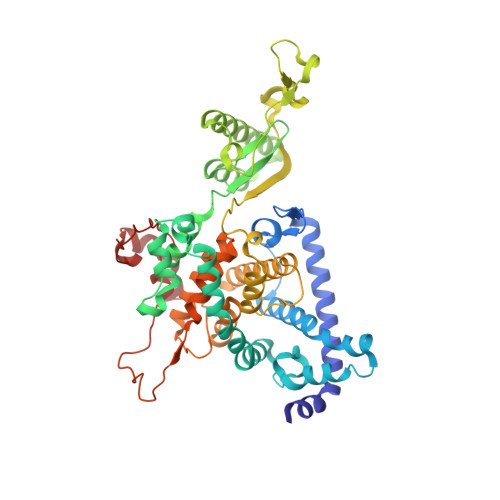
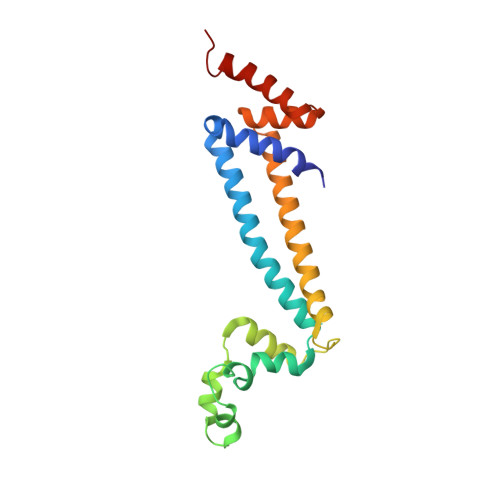
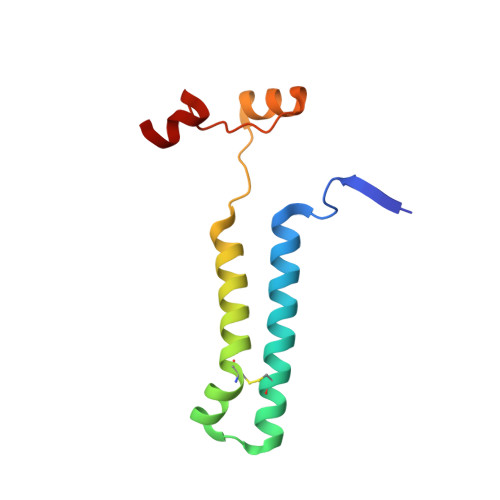
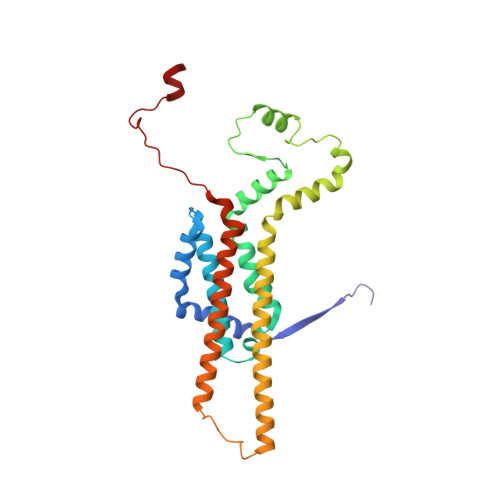
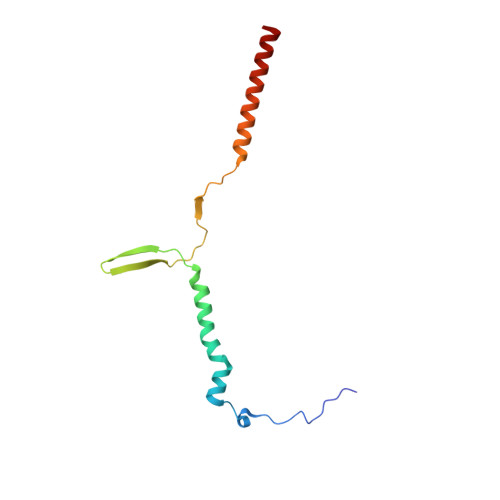
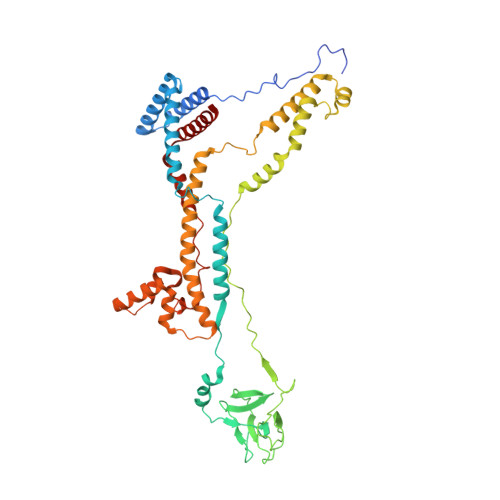
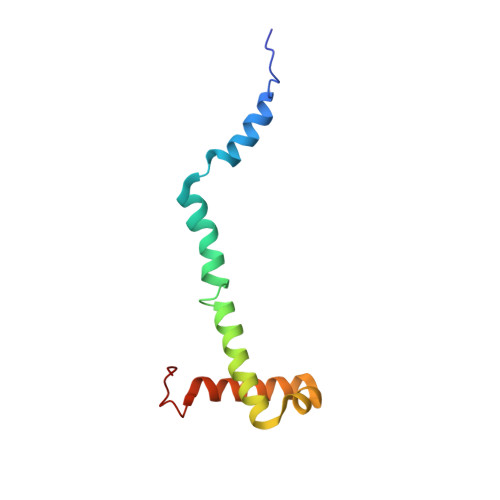
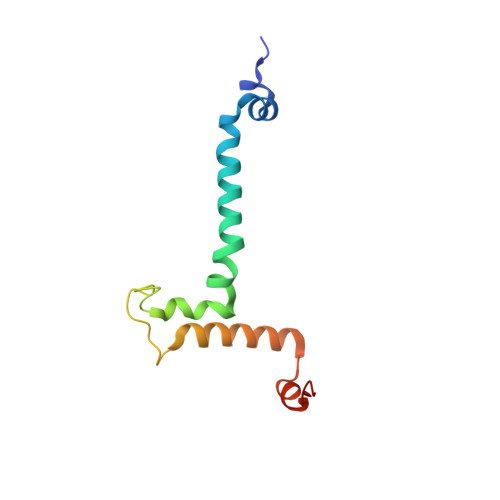
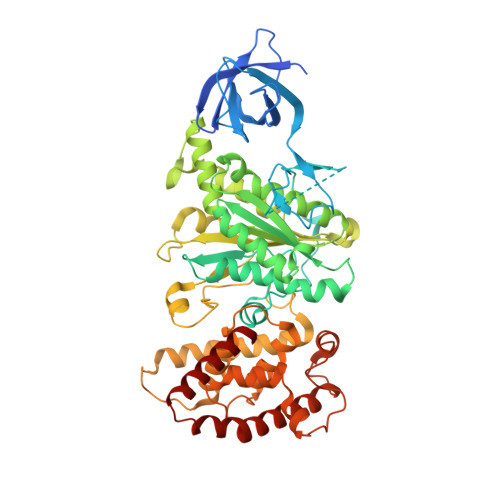
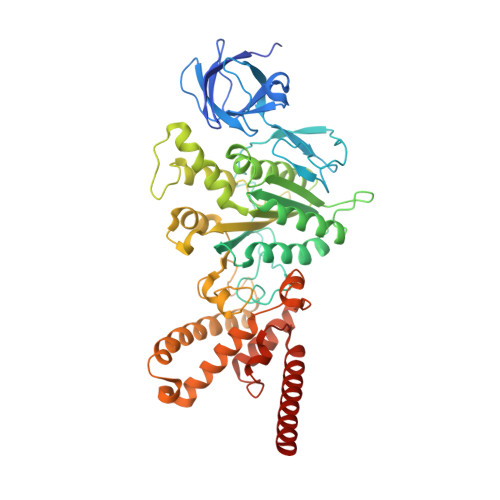
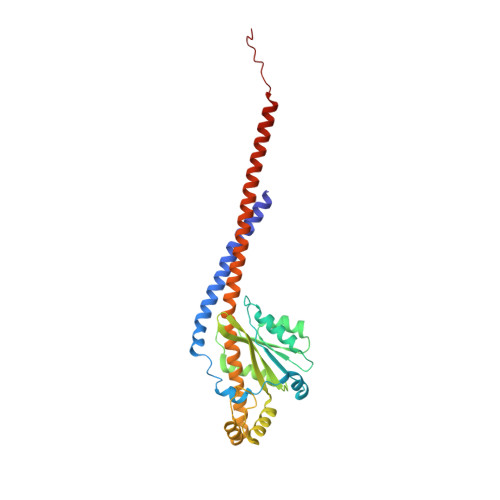
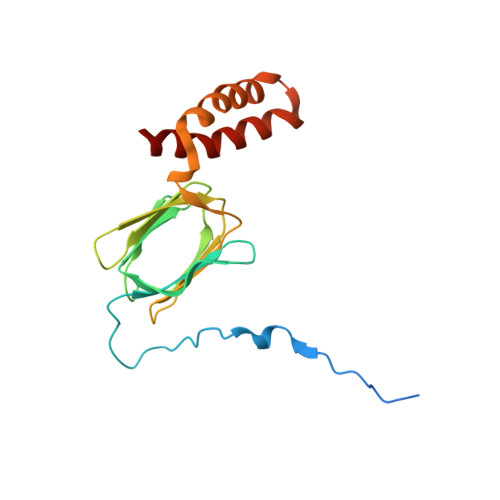
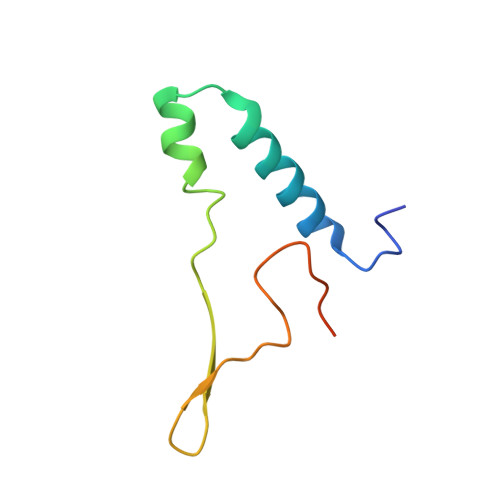
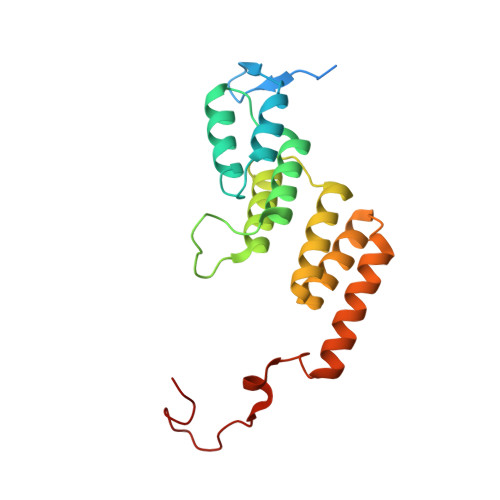
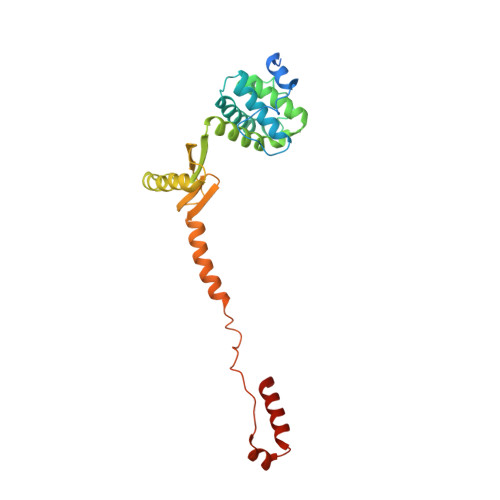
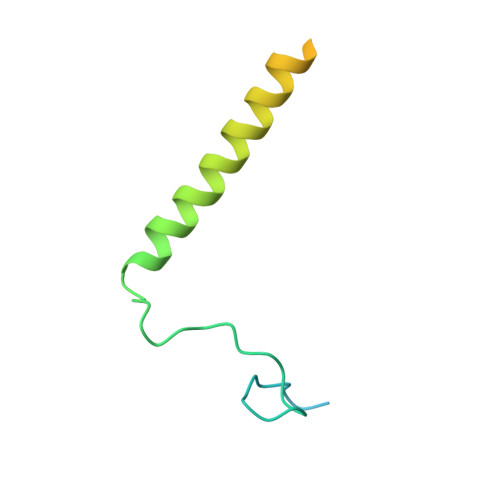
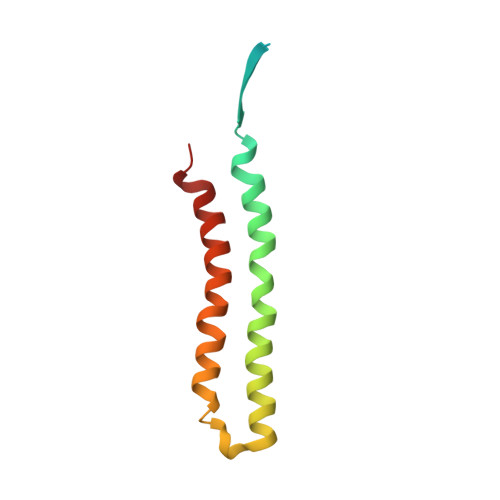
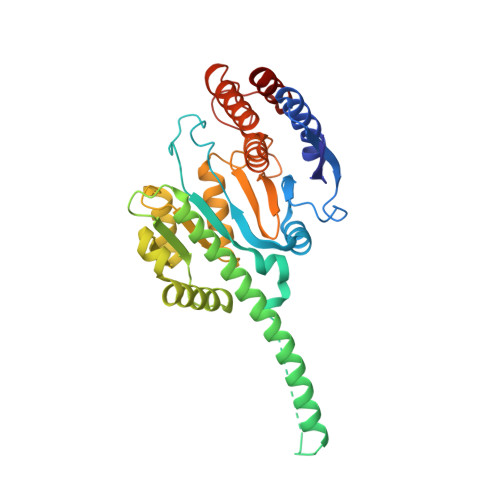
Structure of a mitochondrial ATP synthase with bound native cardiolipin.
Muhleip, A., McComas, S.E., Amunts, A.(2019) Elife 8
- PubMed: 31738165
- DOI: https://doi.org/10.7554/eLife.51179
- Primary Citation of Related Structures:
6TDU, 6TDV, 6TDW, 6TDX, 6TDY, 6TDZ, 6TE0 - PubMed Abstract:
The mitochondrial ATP synthase fuels eukaryotic cells with chemical energy. Here we report the cryo-EM structure of a divergent ATP synthase dimer from mitochondria of Euglena gracilis , a member of the phylum Euglenozoa that also includes human parasites. It features 29 different subunits, 8 of which are newly identified. The membrane region was determined to 2.8 Å resolution, enabling the identification of 37 associated lipids, including 25 cardiolipins, which provides insight into protein-lipid interactions and their functional roles. The rotor-stator interface comprises four membrane-embedded horizontal helices, including a distinct subunit a . The dimer interface is formed entirely by phylum-specific components, and a peripherally associated subcomplex contributes to the membrane curvature. The central and peripheral stalks directly interact with each other. Last, the ATPase inhibitory factor 1 (IF 1 ) binds in a mode that is different from human, but conserved in Trypanosomatids.
- Science for Life Laboratory, Department of Biochemistry and Biophysics, Stockholm University, Solna, Sweden.
Organizational Affiliation: