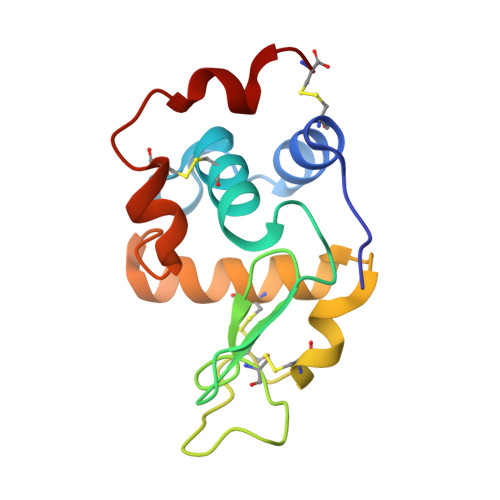
The C-Type Lysozyme from the upper Gastrointestinal Tract of Opisthocomus hoatzin, the Stinkbird.
Taylor, E.J., Skjot, M., Skov, L.K., Klausen, M., De Maria, L., Gippert, G.P., Turkenburg, J.P., Davies, G.J., Wilson, K.S.(2019) Int J Mol Sci 20
- PubMed: 31698762
- DOI: https://doi.org/10.3390/ijms20225531
- Primary Citation of Related Structures:
6T5S, 6T6C - PubMed Abstract:
Muramidases/lysozymes are important bio-molecules, which cleave the glycan backbone in the peptidoglycan polymer found in bacterial cell walls. The glycoside hydrolase (GH) family 22 C-type lysozyme, from the folivorous bird Opisthocomus hoazin (stinkbird), was expressed in Aspergillus oryzae , and a set of variants was produced. All variants were enzymatically active, including those designed to probe key differences between the Hoatzin enzyme and Hen Egg White lysozyme. Four variants showed improved thermostability at pH 4.7, compared to the wild type. The X-ray structure of the enzyme was determined in the apo form and in complex with chitin oligomers. Bioinformatic analysis of avian GH22 amino acid sequences showed that they separate out into three distinct subgroups (chicken-like birds, sea birds and other birds). The Hoatzin is found in the "other birds" group and we propose that this represents a new cluster of avian upper-gut enzymes.
- Structural Biology Laboratory, Department of Chemistry, The University of York, York YO10 5DD, UK.
Organizational Affiliation: