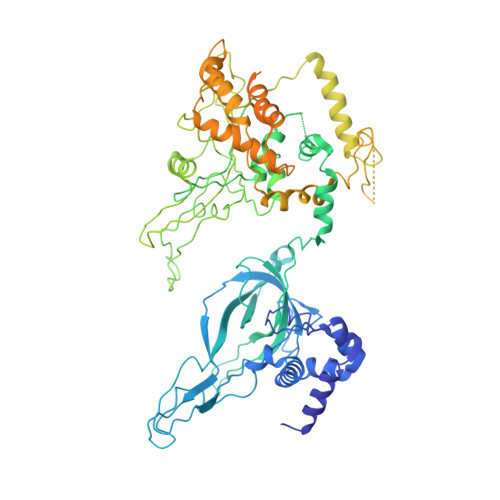
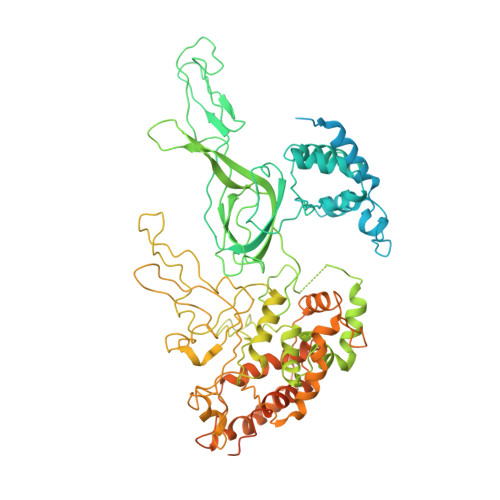
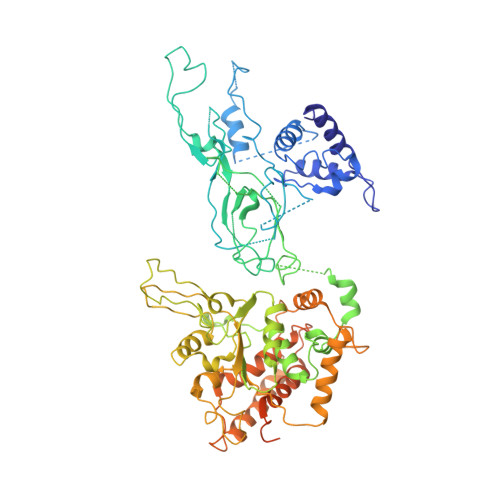
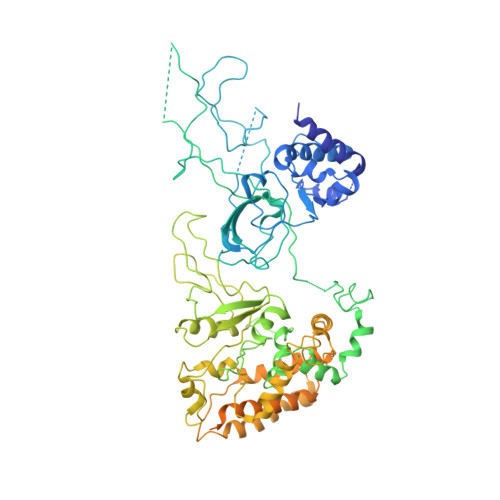
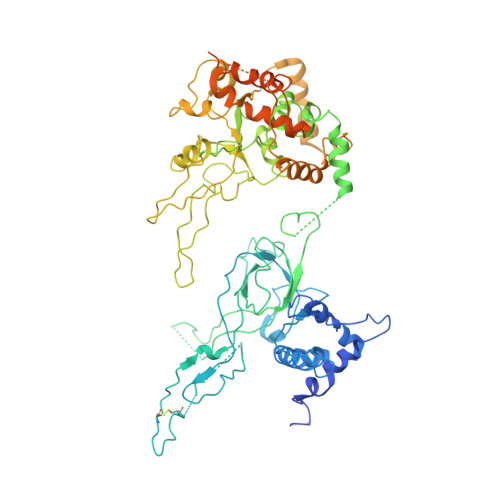
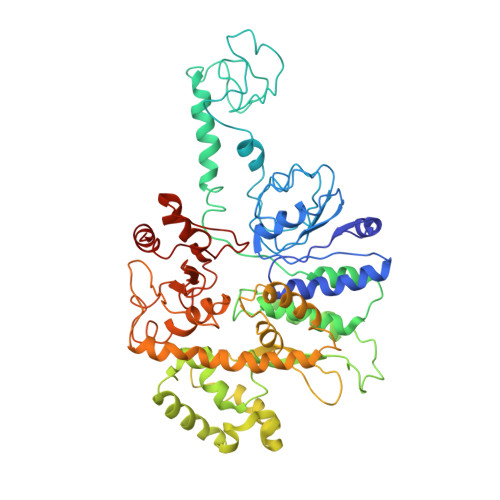
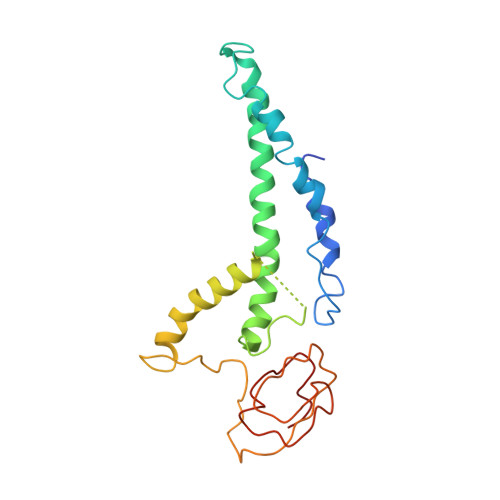
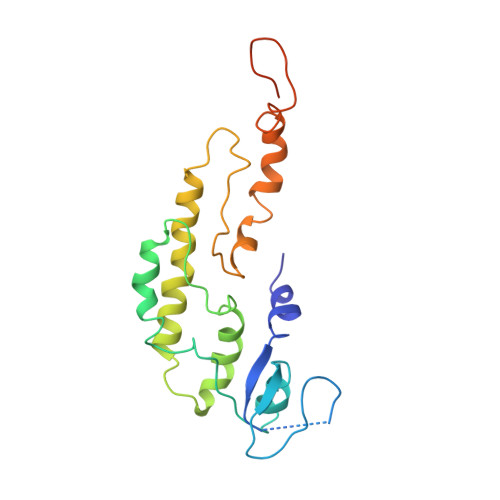
Molecular Basis for ATP-Hydrolysis-Driven DNA Translocation by the CMG Helicase of the Eukaryotic Replisome.
Eickhoff, P., Kose, H.B., Martino, F., Petojevic, T., Abid Ali, F., Locke, J., Tamberg, N., Nans, A., Berger, J.M., Botchan, M.R., Yardimci, H., Costa, A.(2019) Cell Rep 28: 2673-2688.e8
- PubMed: 31484077
- DOI: https://doi.org/10.1016/j.celrep.2019.07.104
- Primary Citation of Related Structures:
6RAW, 6RAX, 6RAY, 6RAZ - PubMed Abstract:
In the eukaryotic replisome, DNA unwinding by the Cdc45-MCM-Go-Ichi-Ni-San (GINS) (CMG) helicase requires a hexameric ring-shaped ATPase named minichromosome maintenance (MCM), which spools single-stranded DNA through its central channel. Not all six ATPase sites are required for unwinding; however, the helicase mechanism is unknown. We imaged ATP-hydrolysis-driven translocation of the CMG using cryo-electron microscopy (cryo-EM) and found that the six MCM subunits engage DNA using four neighboring protomers at a time, with ATP binding promoting DNA engagement. Morphing between different helicase states leads us to suggest a non-symmetric hand-over-hand rotary mechanism, explaining the asymmetric requirements of ATPase function around the MCM ring of the CMG. By imaging of a higher-order replisome assembly, we find that the Mrc1-Csm3-Tof1 fork-stabilization complex strengthens the interaction between parental duplex DNA and the CMG at the fork, which might support the coupling between DNA translocation and fork unwinding.
- Macromolecular Machines Laboratory, The Francis Crick Institute, London NW1 1AT, UK.
Organizational Affiliation: