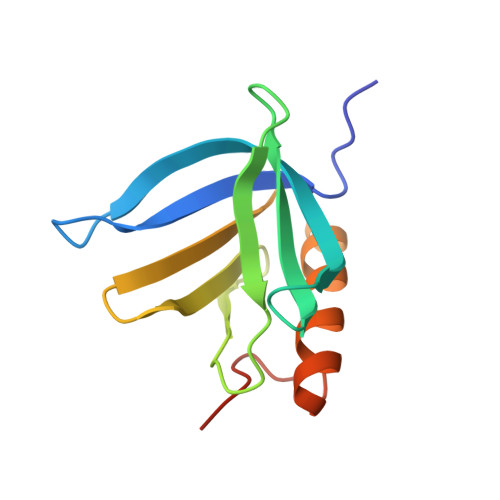
A Consensus Binding Motif for the PP4 Protein Phosphatase.
Ueki, Y., Kruse, T., Weisser, M.B., Sundell, G.N., Larsen, M.S.Y., Mendez, B.L., Jenkins, N.P., Garvanska, D.H., Cressey, L., Zhang, G., Davey, N., Montoya, G., Ivarsson, Y., Kettenbach, A.N., Nilsson, J.(2019) Mol Cell 76: 953
- PubMed: 31585692
- DOI: https://doi.org/10.1016/j.molcel.2019.08.029
- Primary Citation of Related Structures:
6R8I - PubMed Abstract:
Dynamic protein phosphorylation constitutes a fundamental regulatory mechanism in all organisms. Phosphoprotein phosphatase 4 (PP4) is a conserved and essential nuclear serine and threonine phosphatase. Despite the importance of PP4, general principles of substrate selection are unknown, hampering the study of signal regulation by this phosphatase. Here, we identify and thoroughly characterize a general PP4 consensus-binding motif, the FxxP motif. X-ray crystallography studies reveal that FxxP motifs bind to a conserved pocket in the PP4 regulatory subunit PPP4R3. Systems-wide in silico searches integrated with proteomic analysis of PP4 interacting proteins allow us to identify numerous FxxP motifs in proteins controlling a range of fundamental cellular processes. We identify an FxxP motif in the cohesin release factor WAPL and show that this regulates WAPL phosphorylation status and is required for efficient cohesin release. Collectively our work uncovers basic principles of PP4 specificity with broad implications for understanding phosphorylation-mediated signaling in cells.
- Novo Nordisk Foundation Center for Protein Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark.
Organizational Affiliation: