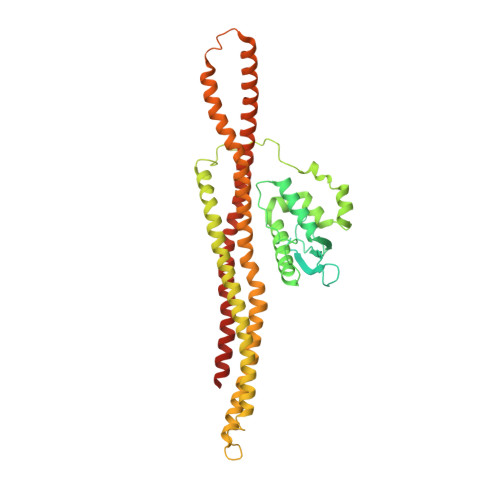
The cryo-EM structure of the SNX-BAR Mvp1 tetramer.
Sun, D., Varlakhanova, N.V., Tornabene, B.A., Ramachandran, R., Zhang, P., Ford, M.G.J.(2020) Nat Commun 11: 1506-1506
- PubMed: 32198400
- DOI: https://doi.org/10.1038/s41467-020-15110-5
- Primary Citation of Related Structures:
6Q0X - PubMed Abstract:
Sorting nexins (SNX) are a family of PX domain-containing proteins with pivotal roles in trafficking and signaling. SNX-BARs, which also have a curvature-generating Bin/Amphiphysin/Rvs (BAR) domain, have membrane-remodeling functions, particularly at the endosome. The minimal PX-BAR module is a dimer mediated by BAR-BAR interactions. Many SNX-BAR proteins, however, additionally have low-complexity N-terminal regions of unknown function. Here, we present the cryo-EM structure of the full-length SNX-BAR Mvp1, which is an autoinhibited tetramer. The tetramer is a dimer of dimers, wherein the membrane-interacting BAR surfaces are sequestered and the PX lipid-binding sites are occluded. The N-terminal low-complexity region of Mvp1 is essential for tetramerization. Mvp1 lacking its N-terminus is dimeric and exhibits enhanced membrane association. Membrane binding and remodeling by Mvp1 therefore requires unmasking of the PX and BAR domain lipid-interacting surfaces. This work reveals a tetrameric configuration of a SNX-BAR protein that provides critical insight into SNX-BAR function and regulation.
- Department of Cell Biology, University of Pittsburgh School of Medicine, Pittsburgh, PA, 15261, USA.
Organizational Affiliation: