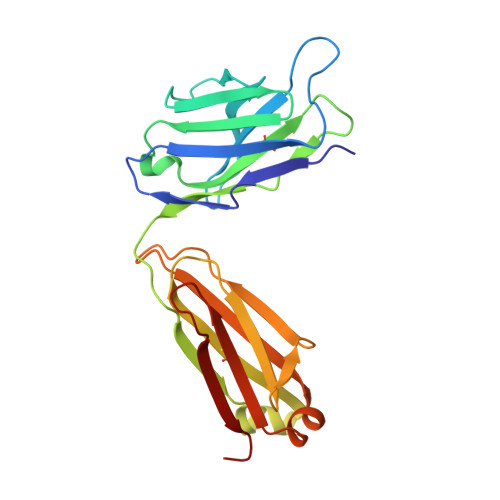
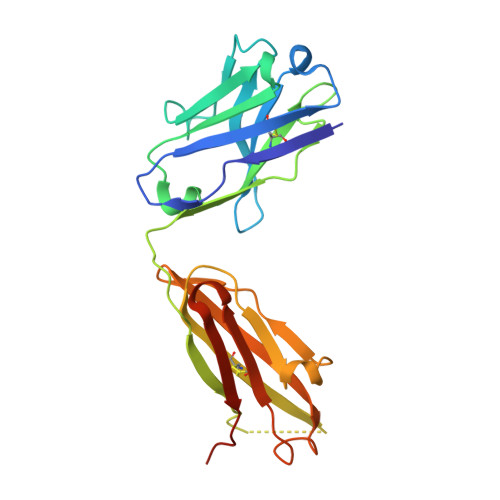
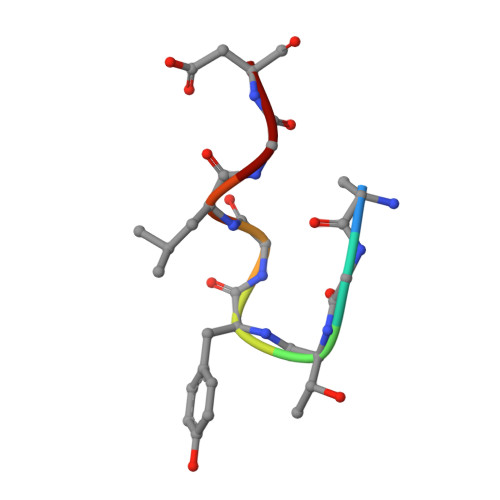
Characterization of tau binding by gosuranemab.
Sopko, R., Golonzhka, O., Arndt, J., Quan, C., Czerkowicz, J., Cameron, A., Smith, B., Murugesan, Y., Gibbons, G., Kim, S.J., Trojanowski, J.Q., Lee, V.M.Y., Brunden, K.R., Graham, D.L., Weinreb, P.H., Hering, H.(2020) Neurobiol Dis 146: 105120-105120
- PubMed: 32991997
- DOI: https://doi.org/10.1016/j.nbd.2020.105120
- Primary Citation of Related Structures:
6PXR - PubMed Abstract:
Deposition of tau aggregates in the brain is a pathological hallmark of several neurodegenerative diseases, termed tauopathies, such as Alzheimer's disease (AD), corticobasal degeneration, and progressive supranuclear palsy (PSP). As transcellular spread of pathological tau aggregates has been implicated in disease progression, immunotherapy is being considered as a treatment for tauopathies. Here we report a detailed biochemical and biophysical characterization of the tau-binding properties of gosuranemab, a humanized monoclonal antibody directed against N-terminal tau that is currently being investigated as a treatment for AD. Binding experiments showed that gosuranemab exhibited high affinity for tau monomer, tau fibrils, and insoluble tau from different tauopathies. Epitope mapping studies conducted using X-ray crystallography and mutagenesis showed that gosuranemab bound to human tau residues 15-22. Immunodepletion of pathological human brain homogenates and transgenic mouse interstitial fluid (ISF) with gosuranemab resulted in reduced tau aggregation in tau biosensor cells. Preincubation of seed-competent AD-tau with gosuranemab significantly inhibited tau aggregation in mouse primary cortical neurons. Gosuranemab also significantly reduced unbound N-terminal tau in cerebrospinal fluid (CSF) from individuals with PSP and AD, and in ISF and CSF of treated transgenic mice. These results are consistent with the >90% target engagement observed in the CSF of some clinical trial dosing cohorts and support the evaluation of gosuranemab as a potential treatment for AD.
- Biogen, Cambridge, MA, USA. Electronic address: richelle.sopko@biogen.com.
Organizational Affiliation: