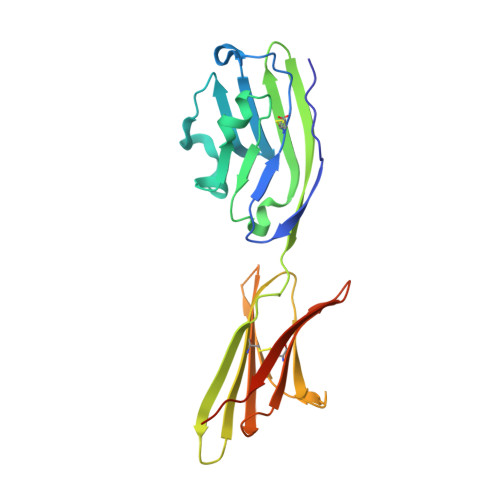
Protein Footprinting and X-ray Crystallography Reveal the Interaction of PD-L1 and a Macrocyclic Peptide.
Niu, B., Appleby, T.C., Wang, R., Morar, M., Voight, J., Villasenor, A.G., Clancy, S., Wise, S., Belzile, J.P., Papalia, G., Wong, M., Brendza, K.M., Lad, L., Gross, M.L.(2020) Biochemistry 59: 541-551
- PubMed: 31841311
- DOI: https://doi.org/10.1021/acs.biochem.9b00822
- Primary Citation of Related Structures:
6PV9 - PubMed Abstract:
Blocking interactions between PD-1 and PD-L1 opens a new era of cancer treatment involving immunity modulation. Although most immunotherapies use monoclonal antibodies, small-molecule inhibitors offer advantages. To facilitate development of small-molecule therapeutics, we implemented a rapid approach to characterize the binding interfaces of small-molecule inhibitors with PD-L1. We determined its interaction with a synthetic macrocyclic peptide by using two mass spectrometry-based approaches, hydrogen-deuterium exchange and fast photochemical oxidation of proteins (FPOP), and corroborated the findings with our X-ray structure of the PD-L1/macrocycle complex. Although all three approaches show that the macrocycle binds directly to PD-L1 over the regions of residues 46-87 and 114-125, the two protein footprinting approaches show additional binding at the N-terminus of PD-L1, and FPOP reveals some critical binding residues. The outcomes not only show the binding regions but also demonstrate the utility of MS-based footprinting in probing protein/ligand inhibitory interactions in cancer immunotherapy.
- Department of Chemistry , Washington University in St. Louis , St. Louis , Missouri 63130 , United States.
Organizational Affiliation: