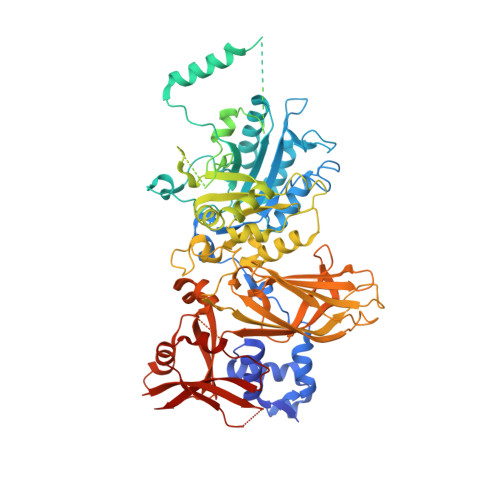
Structure of phospholipase C epsilon reveals an integrated RA1 domain and previously unidentified regulatory elements.
Rugema, N.Y., Garland-Kuntz, E.E., Sieng, M., Muralidharan, K., Van Camp, M.M., O'Neill, H., Mbongo, W., Selvia, A.F., Marti, A.T., Everly, A., McKenzie, E., Lyon, A.M.(2020) Commun Biol 3: 445-445
- PubMed: 32796910
- DOI: https://doi.org/10.1038/s42003-020-01178-8
- Primary Citation of Related Structures:
6PMP - PubMed Abstract:
Phospholipase Cε (PLCε) generates lipid-derived second messengers at the plasma and perinuclear membranes in the cardiovascular system. It is activated in response to a wide variety of signals, such as those conveyed by Rap1A and Ras, through a mechanism that involves its C-terminal Ras association (RA) domains (RA1 and RA2). However, the complexity and size of PLCε has hindered its structural and functional analysis. Herein, we report the 2.7 Å crystal structure of the minimal fragment of PLCε that retains basal activity. This structure includes the RA1 domain, which forms extensive interactions with other core domains. A conserved amphipathic helix in the autoregulatory X-Y linker of PLCε is also revealed, which we show modulates activity in vitro and in cells. The studies provide the structural framework for the core of this critical cardiovascular enzyme that will allow for a better understanding of its regulation and roles in disease.
- Department of Chemistry, Purdue University, West Lafayette, 47907, IN, USA.
Organizational Affiliation: