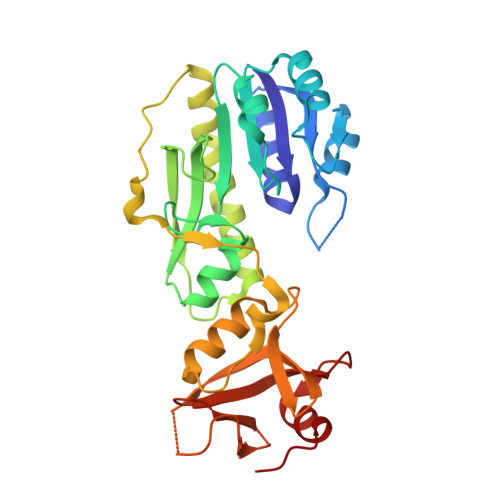
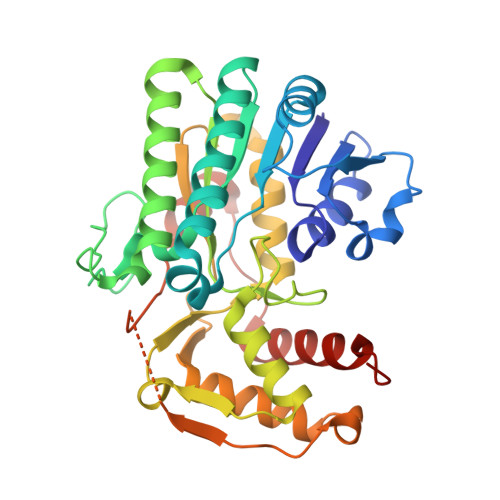
Cryo-electron microscopy structures of ArnA, a key enzyme for polymyxin resistance, revealed unexpected oligomerizations and domain movements.
Yang, M., Chen, Y.S., Ichikawa, M., Calles-Garcia, D., Basu, K., Fakih, R., Bui, K.H., Gehring, K.(2019) J Struct Biol 208: 43-50
- PubMed: 31344437
- DOI: https://doi.org/10.1016/j.jsb.2019.07.009
- Primary Citation of Related Structures:
6PIH, 6PIK - PubMed Abstract:
Gram-negative bacteria evade the attack of cationic antimicrobial peptides through modifying their lipid A structure in their outer membranes with 4-amino-4-deoxy-L-arabinose (Ara4N). ArnA is a crucial enzyme in the lipid A modification pathway and its deletion abolishes the polymyxin resistance of gram-negative bacteria. Previous studies by X-ray crystallography have shown that full-length ArnA forms a three-bladed propeller-shaped hexamer. Here, the structures of ArnA determined by cryo-electron microscopy (cryo-EM) reveal that ArnA exists in two 3D architectures, hexamer and tetramer. This is the first observation of a tetrameric ArnA. The hexameric cryo-EM structure is similar to previous crystal structures but shows differences in domain movements and conformational changes. We propose that ArnA oligomeric states are in a dynamic equilibrium, where the hexamer state is energetically more favorable, and its domain movements are important for cooperating with downstream enzymes in the lipid A-Ara4N modification pathway. The results provide us with new possibilities to explore inhibitors targeting ArnA.
- Department of Biochemistry & Centre for Structural Biology, McGill University, Montreal, QC H3G 0B1, Canada. Electronic address: meng.yang@mcgill.ca.
Organizational Affiliation: