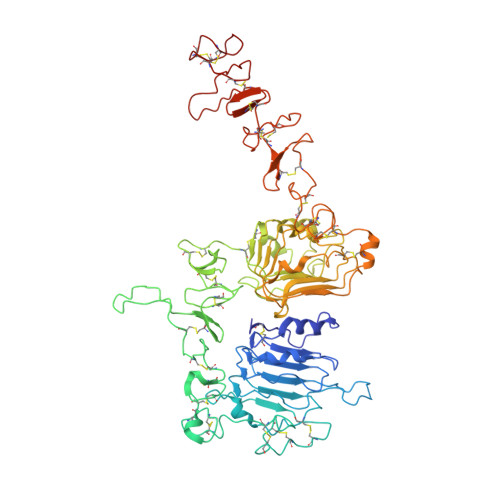
Cryo-EM Structure of HER2-trastuzumab-pertuzumab complex.
Hao, Y., Yu, X., Bai, Y., McBride, H.J., Huang, X.(2019) PLoS One 14: e0216095-e0216095
- PubMed: 31042744
- DOI: https://doi.org/10.1371/journal.pone.0216095
- Primary Citation of Related Structures:
6OGE - PubMed Abstract:
Trastuzumab and pertuzumab are monoclonal antibodies that bind to distinct subdomains of the extracellular domain of human epidermal growth factor receptor 2 (HER2). Adding these monoclonal antibodies to the treatment regimen of HER2-positive breast cancer has changed the paradigm for treatment in that form of cancer. Synergistic activity has been observed with the combination of these two antibodies leading to hypotheses regarding the mechanism(s) and to the development of bispecific antibodies to maximize the clinical effect further. Although the individual crystal structures of HER2-trastuzumab and HER2-pertuzumab revealed the distinct binding sites and provided the structural basis for their anti-tumor activities, detailed structural information on the HER2-trastuzumab-pertuzumab complex has been elusive. Here we present the cryo-EM structure of HER2-trastuzumab-pertuzumab at 4.36 Å resolution. Comparison with the binary complexes reveals no cooperative interaction between trastuzumab and pertuzumab, and provides key insights into the design of novel, high-avidity bispecific molecules with potentially greater clinical efficacy.
- Department of Molecular Engineering, Amgen Inc., Cambridge, MA, United States of America.
Organizational Affiliation: