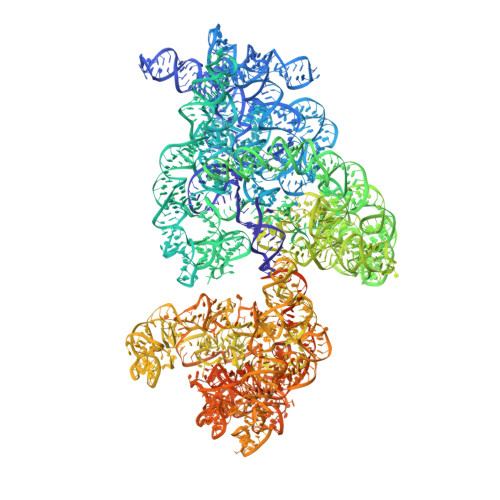
Role of Era in assembly and homeostasis of the ribosomal small subunit.
Razi, A., Davis, J.H., Hao, Y., Jahagirdar, D., Thurlow, B., Basu, K., Jain, N., Gomez-Blanco, J., Britton, R.A., Vargas, J., Guarne, A., Woodson, S.A., Williamson, J.R., Ortega, J.(2019) Nucleic Acids Res 47: 8301-8317
- PubMed: 31265110
- DOI: https://doi.org/10.1093/nar/gkz571
- Primary Citation of Related Structures:
6NQB - PubMed Abstract:
Assembly factors provide speed and directionality to the maturation process of the 30S subunit in bacteria. To gain a more precise understanding of how these proteins mediate 30S maturation, it is important to expand on studies of 30S assembly intermediates purified from bacterial strains lacking particular maturation factors. To reveal the role of the essential protein Era in the assembly of the 30S ribosomal subunit, we analyzed assembly intermediates that accumulated in Era-depleted Escherichia coli cells using quantitative mass spectrometry, high resolution cryo-electron microscopy and in-cell footprinting. Our combined approach allowed for visualization of the small subunit as it assembled and revealed that with the exception of key helices in the platform domain, all other 16S rRNA domains fold even in the absence of Era. Notably, the maturing particles did not stall while waiting for the platform domain to mature and instead re-routed their folding pathway to enable concerted maturation of other structural motifs spanning multiple rRNA domains. We also found that binding of Era to the mature 30S subunit destabilized helix 44 and the decoding center preventing binding of YjeQ, another assembly factor. This work establishes Era's role in ribosome assembly and suggests new roles in maintaining ribosome homeostasis.
- Department of Anatomy and Cell Biology, McGill University, Montreal, Quebec H3A 0C7, Canada.
Organizational Affiliation: