Structure, Function, and Dynamics of the G alpha Binding Domain of Ric-8A.
Zeng, B., Mou, T.C., Doukov, T.I., Steiner, A., Yu, W., Papasergi-Scott, M., Tall, G.G., Hagn, F., Sprang, S.R.(2019) Structure 27: 1137-1147.e5
- PubMed: 31155309
- DOI: https://doi.org/10.1016/j.str.2019.04.013
- Primary Citation of Related Structures:
6NMG, 6NMJ - PubMed Abstract:
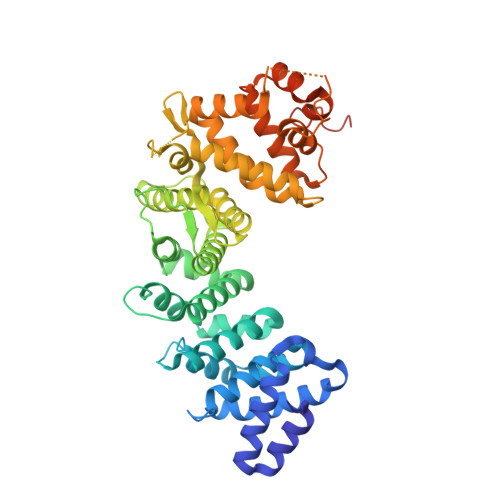
Ric-8A is a 530-amino acid cytoplasmic molecular chaperone and guanine nucleotide exchange factor (GEF) for i, q, and 12/13 classes of heterortrimeric G protein alpha subunits (Gα). We report the 2.2-Å crystal structure of the Ric-8A Gα-binding domain with GEF activity, residues 1-452, and is phosphorylated at Ser435 and Thr440. Residues 1-429 adopt a superhelical fold comprised of Armadillo (ARM) and HEAT repeats, and the C terminus is disordered. One of the phosphorylated residues potentially binds to a basic cluster in an ARM motif. Amino acid sequence conservation and published hydrogen-deuterium exchange data indicate repeats 3 through 6 to be a putative Gα-binding surface. Normal mode modeling of small-angle X-ray scattering data indicates that phosphorylation induces relative rotation between repeats 1-4, 5-6, and 7-9. 2D 1 H- 15 N-TROSY spectra of [ 2 H, 15 N]-labeled Gαi1 in the presence of R452 reveals chemical shift perturbations of the C terminus and Gαi1 residues involved in nucleotide binding.
- Graduate Program in Biochemistry and Biophysics, University of Montana, Missoula, MT 59812, USA.
Organizational Affiliation: