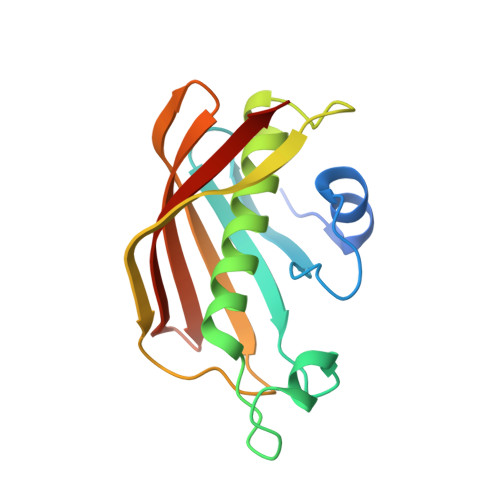
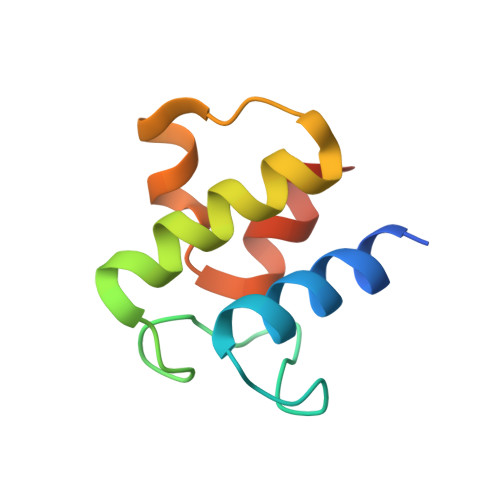
Structural and dynamical rationale for fatty acid unsaturation inEscherichia coli.
Dodge, G.J., Patel, A., Jaremko, K.L., McCammon, J.A., Smith, J.L., Burkart, M.D.(2019) Proc Natl Acad Sci U S A 116: 6775-6783
- PubMed: 30872475
- DOI: https://doi.org/10.1073/pnas.1818686116
- Primary Citation of Related Structures:
6N3P - PubMed Abstract:
Fatty acid biosynthesis in α- and γ-proteobacteria requires two functionally distinct dehydratases, FabA and FabZ. Here, mechanistic cross-linking facilitates the structural characterization of a stable hexameric complex of six Escherichia coli FabZ dehydratase subunits with six AcpP acyl carrier proteins. The crystal structure sheds light on the divergent substrate selectivity of FabA and FabZ by revealing distinct architectures of the binding pocket. Molecular dynamics simulations demonstrate differential biasing of substrate orientations and conformations within the active sites of FabA and FabZ such that FabZ is preorganized to catalyze only dehydration, while FabA is primed for both dehydration and isomerization.
- Department of Biological Chemistry, University of Michigan, Ann Arbor, MI 48109-2216.
Organizational Affiliation: