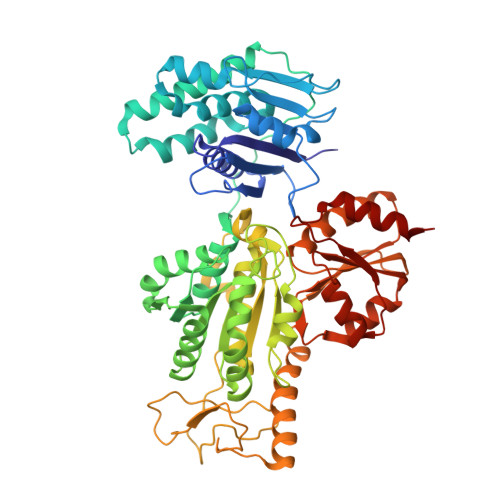
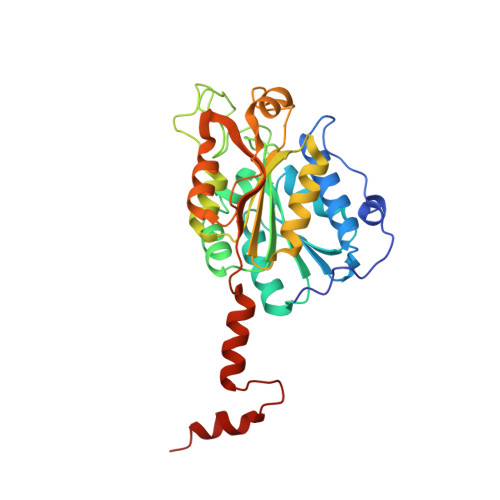
A reverse TCA cycle 2-oxoacid:ferredoxin oxidoreductase that makes C-C bonds from CO2.
Chen, P.Y., Li, B., Drennan, C.L., Elliott, S.J.(2019) Joule 3: 595-611
- PubMed: 31080943
- DOI: https://doi.org/10.1016/j.joule.2018.12.006
- Primary Citation of Related Structures:
6N2N, 6N2O - PubMed Abstract:
2-oxoglutarate:ferredoxin oxidoreductase (OGOR) is a thiamine pyrophosphate (TPP) and [4Fe-4S] cluster-dependent enzyme from the reductive tricarboxylic acid (rTCA) cycle that fixes CO 2 to succinyl-CoA, forming 2-oxoglutarate and CoA. Here we report an OGOR from the rTCA cycle of Magnetococcus marinus MC-1, along with all three potential ferredoxin (Fd) redox partners. We demonstrate Mm OGOR operates bidirectionally (both CO 2 -fixing and 2-oxoglutarate oxidizing), and that only one Fd ( Mm Fd1) supports efficient catalysis. Our 1.94-Å and 2.80-Å resolution crystal structures of native and substrate-bound forms of Mm OGOR reveal the determinants of substrate specificity and CoA-binding in an OGOR, and illuminate the [4Fe-4S] cluster environment, portraying the electronic conduit allowing Mm Fd1 to be wired to the bound-TPP. Structural and biochemical data further identify Glu45α as a mobile residue that impacts catalytic bias toward CO 2 -fixation although it makes no direct contact with TPP-bound intermediates, indicating that reaction directionality can be tuned by second layer interactions. (149 of 150 words limit).
- Department of Chemistry, Massachusetts Institute of Technology, Cambridge, MA 02139.
Organizational Affiliation: