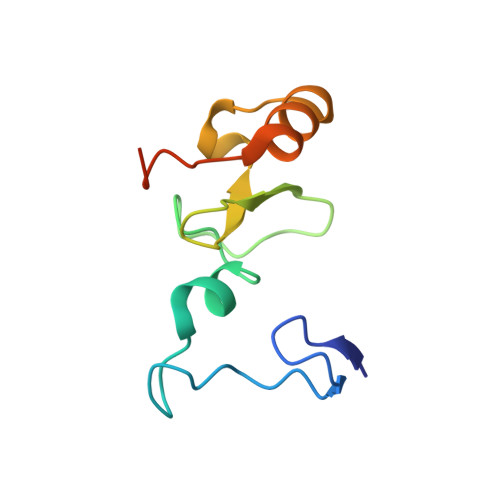
Structural insights into trans-histone regulation of H3K4 methylation by unique histone H4 binding of MLL3/4.
Liu, Y., Qin, S., Chen, T.Y., Lei, M., Dhar, S.S., Ho, J.C., Dong, A., Loppnau, P., Li, Y., Lee, M.G., Min, J.(2019) Nat Commun 10: 36-36
- PubMed: 30604749
- DOI: https://doi.org/10.1038/s41467-018-07906-3
- Primary Citation of Related Structures:
6MLC - PubMed Abstract:
MLL3 and MLL4 are two closely related members of the SET1/MLL family of histone H3K4 methyltransferases and are responsible for monomethylating histone H3K4 on enhancers, which are essential in regulating cell-type-specific gene expression. Mutations of MLL3 or MLL4 have been reported in different types of cancer. Recently, the PHD domains of MLL3/4 have been reported to recruit the MLL3/4 complexes to their target genes by binding to histone H4 during the NT2/D1 stem cell differentiation. Here we show that an extended PHD domain (ePHD 6 ) involving the sixth PHD domain and its preceding zinc finger in MLL3 and MLL4 specifically recognizes an H4H18-containing histone H4 fragment and that modifications of residues surrounding H4H18 modulate H4 binding to MLL3/4. Our in vitro methyltransferase assays and cellular experiments further reveal that the interaction between ePHD 6 of MLL3/4 and histone H4 is required for their nucleosomal methylation activity and MLL4-mediated neuronal differentiation of NT2/D1 cells.
- Structural Genomics Consortium, University of Toronto, 101 College Street, Toronto, Ontario, M5G 1L7, Canada.
Organizational Affiliation: