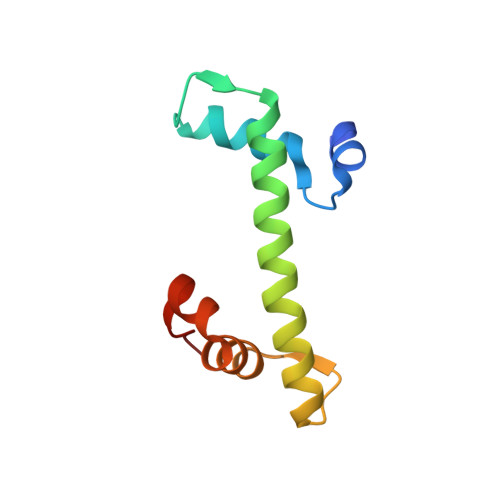
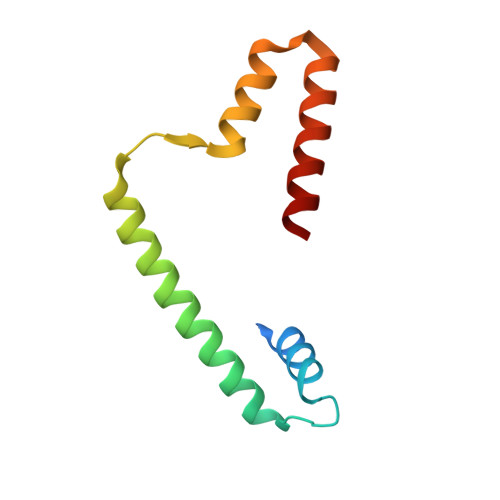
OsChz1 acts as a histone chaperone in modulating chromatin organization and genome function in rice.
Du, K., Luo, Q., Yin, L., Wu, J., Liu, Y., Gan, J., Dong, A., Shen, W.H.(2020) Nat Commun 11: 5717-5717
- PubMed: 33177521
- DOI: https://doi.org/10.1038/s41467-020-19586-z
- Primary Citation of Related Structures:
6M2M - PubMed Abstract:
While the yeast Chz1 acts as a specific histone-chaperone for H2A.Z, functions of CHZ-domain proteins in multicellular eukaryotes remain obscure. Here, we report on the functional characterization of OsChz1, a sole CHZ-domain protein identified in rice. OsChz1 interacts with both the canonical H2A-H2B dimer and the variant H2A.Z-H2B dimer. Within crystal structure the C-terminal region of OsChz1 binds H2A-H2B via an acidic region, pointing to a previously unknown recognition mechanism. Knockout of OsChz1 leads to multiple plant developmental defects. At genome-wide level, loss of OsChz1 causes mis-regulations of thousands of genes and broad alterations of nucleosome occupancy as well as reductions of H2A.Z-enrichment. While OsChz1 associates with chromatin regions enriched of repressive histone marks (H3K27me3 and H3K4me2), its loss does not affect the genome landscape of DNA methylation. Taken together, it is emerging that OsChz1 functions as an important H2A/H2A.Z-H2B chaperone in dynamic regulation of chromatin for higher eukaryote development.
- State Key Laboratory of Genetic Engineering, Collaborative Innovation Center of Genetics and Development, International Associated Laboratory of CNRS-Fudan-HUNAU on Plant Epigenome Research, Department of Biochemistry, Institute of Plant Biology, School of Life Sciences, Fudan University, Shanghai, 200438, China.
Organizational Affiliation: