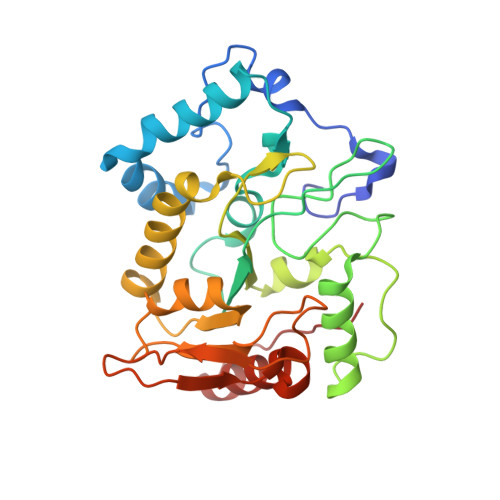
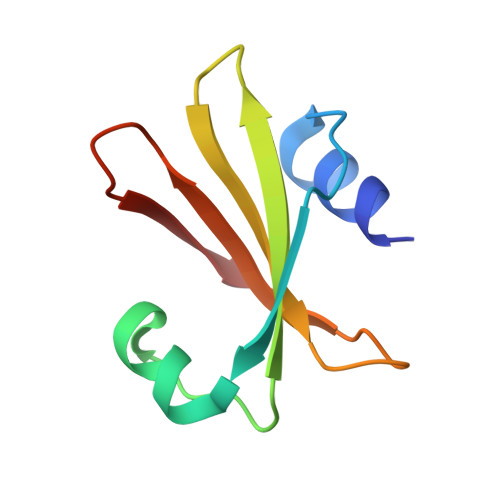
Selective interactions between mimivirus uracil-DNA glycosylase and inhibitory proteins determined by a single amino acid.
Pathak, D., Kwon, E., Kim, D.Y.(2020) J Struct Biol 211: 107552-107552
- PubMed: 32569642
- DOI: https://doi.org/10.1016/j.jsb.2020.107552
- Primary Citation of Related Structures:
6LYD, 6LYE - PubMed Abstract:
Uracil-N-glycosylase (UNG) is found in most organisms as well as in large DNA viruses. Its inhibitory proteins, including uracil glycosylase inhibitor (UGI) and p56, tightly bind to the active site of UNG by mimicking the DNA substrates. As the binding motifs are conserved in UNG family proteins, the inhibitory proteins bind to various UNG proteins across species. However, the intercalation residue that penetrates the DNA minor groove during uracil excision is not conserved among UNG proteins. To understand the role of the intercalation residue in their binding to the inhibitory proteins, we prepared mutants of mimivirus UNG, measured the binding affinity between the UNG mutants and inhibitory proteins, and analyzed the interactions based on the crystal structures of mimivirus UNG mutants complexed with UGI. The results show that mimivirus UNG, which harbors Tyr as an intercalation residue, did not interact with the inhibitory proteins intrinsically, whereas mutations of the intercalation residue to Phe or Leu resulted in tight interactions with UGI and p56; mutation to Met resulted in tight interactions only with p56. The crystal structures revealed that Phe and Leu stabilize the interactions by fitting into the hydrophobic pocket of UGI. These results show that differences in size and hydrophobicity of the intercalation residues determine the interactions between UNG family proteins and the inhibitory proteins, UGI and p56.
- College of Pharmacy, Yeungnam University, Gyeongsan, Gyeongbuk 38541, South Korea.
Organizational Affiliation: